Figures & data
Figure 1. The Sample picture of C. fordii. The picture taken by Huang Yonglin in the Haiyang Mountain Nature Reserve.

Figure 2. The Genome map of C. fordii. Genes inside the circle are transcribed clockwise, and those on the outside are transcribed counter-clockwise. Genes are colored according to functional categories. The darker grey area in the inner circle corresponds to GC content, whereas the lighter grey corresponds to AT content.
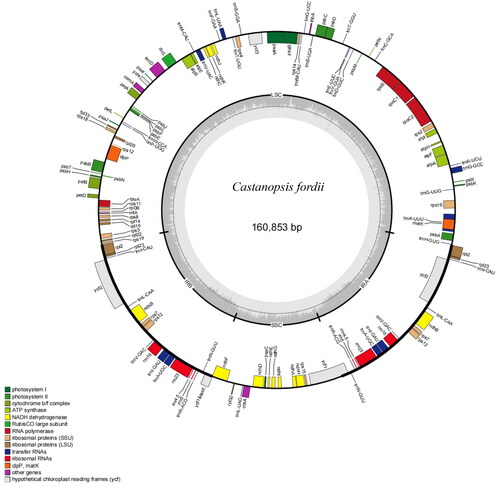
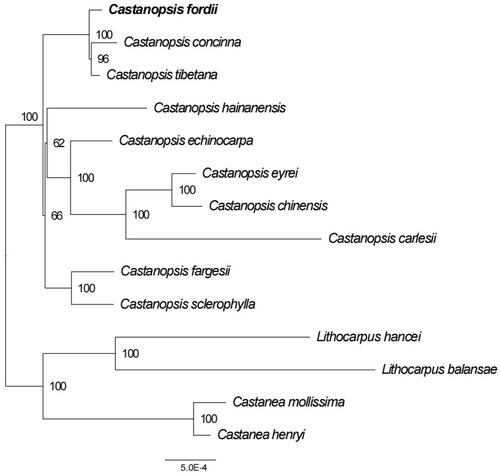
Figure 3. The ML phylogenetic tree based on fourteen complete chloroplast genomes. Maximum-likelihood phylogenetic tree was constructed by GTRCATI Model of RAxML based on complete cp genomes. The sequence type is the nucleotide sequence of the conservative coding gene. The bootstrap check value is 100. Numbers close to each node are bootstrap support values. Accession number: Castanopsis concinna KT793041.1 (Shi et al. Citation2022), Castanopsis hainanensis MG383644.1 (Ogoma et al. Citation2022), Castanopsis fargesii NC_047230.1 (Li et al. Citation2022), Castanopsis carlesii NC_057119.1 (Wang et al. Citation2022), Castanopsis sclerophylla MT627605.1 (Miao et al. Citation2020), Castanopsis echinocarpa NC_023801.1 (Guo et al. Citation2022), Castanopsis tibetana ON710842 (Sanchez-Puerta and Abbona Citation2014), Castanopsis eyrei ON550006, Castanopsis chinensis ON500675, Castanopsis fordii ON710841 (Li et al. Citation2022), Lithocarpus balansae KP299291.1 (Shi et al. Citation2022), Castanea henryi KX954615.1 (Shi et al. Citation2022), Castanea mollissima NC_014674 (Jansen et al. Citation2011), Lithocarpus hancei MW375417.1 (Shi et al. Citation2022). Castanopsis eyrei and Castanopsis chinensis have no publications and can only cite the sequences by accession numbers.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. ON710841. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA864116, SRR20726142, and SAMN30074217 respectively.”
