Figures & data
Figure 1. The morphological characteristics of R. irritans. Photos A and B show the entire plant and its growing environment, respectively (photos taken by Shibing Yang in General Ditch, Qinghai Province).

Figure 2. Genome map of R. irritans. The forward-encoding and reverse-encoding genes are located on the inner and outer sides of the circle, respectively.
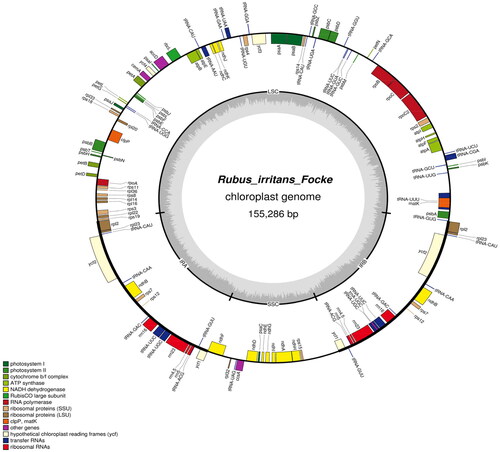
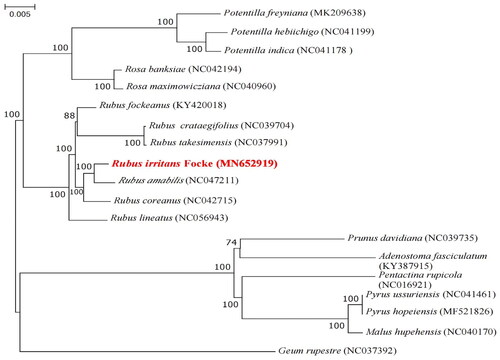
Figure 3. Phylogenomic tree of R. irritans (in red) and 18 species constructed using ML method based on complete cp genome sequences. Numbers in the nodes were bootstrap values from 1000 replicates. The following sequences were used: Potentilla freyniana (Park et al. Citation2019), Potentilla hebiichigo, Potentilla indica (Liu et al. Citation2021), Rosa banksiae, Rosa maximowicziana (Kim et al. Citation2019), Rubus fockeanus, Rubus crataegifolius, Rubus takesimensis (Liu et al. Citation2021), Rubus amabilis, Rubus coreanus (Zhang et al. Citation2021), Rubus lineatus (Lim et al. Citation2021), Prunus davidiana (Wang et al. Citation2020), Adenostoma fasciculatum KY387915 (Kim and Kim Citation2016), Pyrus ussuriensis, Pyrus hopeiensis (Cho et al. Citation2019), Malus hupehensis (Li et al. Citation2020), Geum rupestre (Zhang et al. Citation2022).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. MN652919. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA771538, SRR16529182, and SAMN22315382, respectively.
