Figures & data
Figure 1. The morphological characteristics of Dracocephalum rupestre. D. rupestre is a perennial, 20–30 cm tall. Stems ascending, unbranched, pubescent; leaf blade cordiform or reinform, crenate, white villous; corolla purple-blue, pubescent. The photo was taken by Dr. Tae-Im Heo in Mt. Deokhang, South Korea, June 2022, without any copyright issues.

Figure 2. Schematic map of overall features of the chloroplast genome of Dracocephalum rupestre. The circular map of the chloroplast genome was generated using CPGview (Liu et al. Citation2023). The map contains seven circles. From the center going outward, the first circle shows the distributed repeats connected with red (the forward direction) and green (the reverse direction) arcs. The next circle shows the tandem repeats marked with short bars. The third circle shows the microsatellite sequences as short bars. The fourth circle shows the size of the LSC and SSC. The fifth circle shows the IRA and IRB. The sixth circle shows the GC contents along the plastome. The seventh circle shows the genes having different colors based on their functional groups.
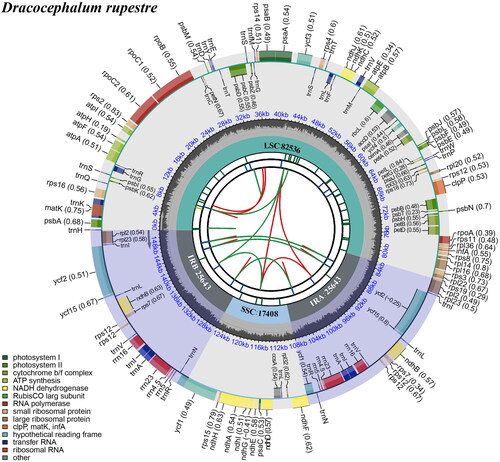
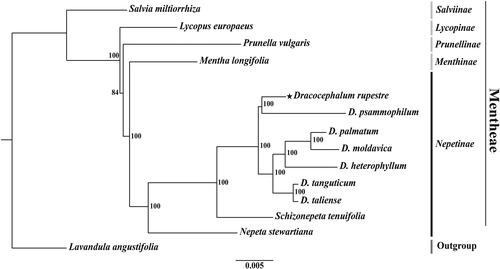
Figure 3. A phylogenetic tree (RAxML) was established based on concatenated sequences of 75 common protein-coding genes in 14 complete chloroplast genomes of the family Lamiaceae. The sequences used for tree construction are as follows: Salvia miltiorrhiza (JX312195; Qian et al. Citation2013), Lycopus europaeus (OM617843), Prunella vulgaris (MZ636547), Mentha longifolia (KU956042; Vining et al. Citation2017), Dracocephalum rupestre (OP526971), D. psammophilum (MZ750980), D. palmatum (KU958581), D. moldavica (MT457747; Yao et al. Citation2020), D. heterophyllum (MW970109; Zhang et al. Citation2021), D. tanguticum (MT457746; Yao et al. Citation2020), D. taliense (MT473756; Zhao et al. Citation2021), Schizonepeta tenuifolia (MW900176), Nepeta stewartiana (MT733874; Wu et al. Citation2021), Lavandula angustifolia (KT948988). The numbers above the nodes indicate bootstrap values with 1,000 replicates.
Supplemental Material
Download JPEG Image (448.9 KB)Supplemental Material
Download JPEG Image (41.7 KB)Supplemental Material
Download MS Word (328.9 KB)Data availability statement
Data from this study are available in the NCBI GenBank at https://www.ncbi.nlm.nih.gov (accession number OP526971). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA883819, SRS15224747, and SAMN30996907, respectively.
