Figures & data
Figure 1. Morphological characteristics of Juglans mandshurica. (a) whole plant, (b) fruits, (c) inflorescence, (d) seeds, (e) leaves.

Figure 2. J. mandshurica mitochondrial (mt) genome gene map. The gene map denotes annotated genes based on different functional groups, which are color-coded on the outer circle as transcribed clock-wise (outside) and counter clock-wise (inside). The inner circle indicates the GC content as a dark grey plot.
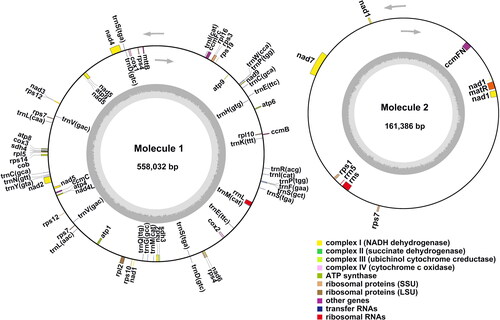
Table 1. Gene composition of the J. mandshurica mt genome.
Supplemental Material
Download MS Word (73.5 KB)Data availability statement
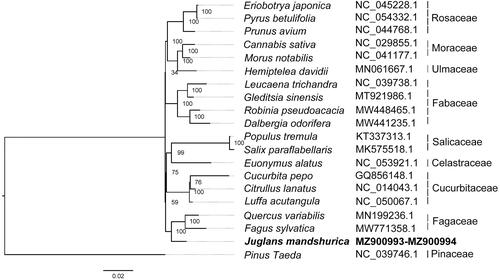
The genome sequence data are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ900993 and MZ900994. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA757787, SRR15615977, SRR15615978, and SAMN20981794 respectively.