Figures & data
Figure 1. Picture of Glandora prostrata subsp. lusitanica isolate BPTPS049 on the day of its collection from a wild population in the dunes of the Beja municipality (Vila Nova de Mil Fontes) in Portugal (collection date: 19 March 2019; location: 37.71500 N, 8.78361 W). BioSample: SAMN28118496.

Figure 2. Graphical map of the complete chloroplast of Glandora prostrata subsp. lusitanica isolate BPTPS049 based on the conversion of annotations openly available in GenBank (accession number: ON641304), color coded based on their functional group, using OrganellarGenomeDRAW (OGDRAW) version 1.3.1 (Greiner et al. Citation2019). Genes inside the circle are transcribed clockwise, genes outside the circle counterclockwise, and intron-containing genes are marked by an asterisk (*). LSC: large single-copy region; SSC: small single-copy region; IRA, IRB: inverted repeats. The dark grey inner ring represents the GC content, while the complementary light grey ring represents the AT content.
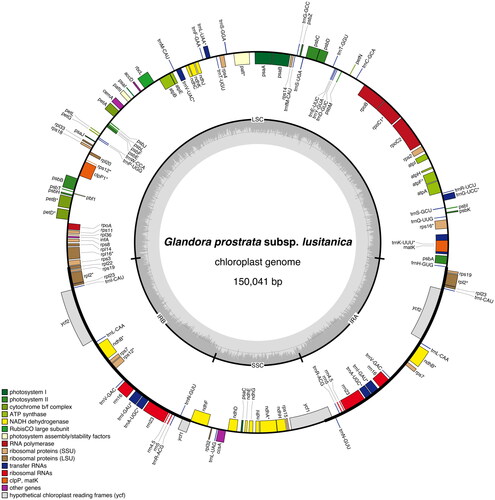
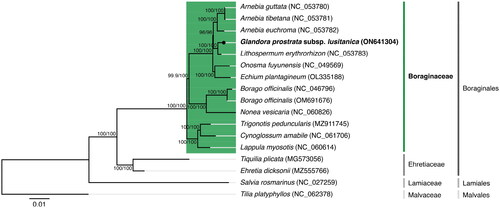
Figure 3. Maximum-likelihood tree inferred from the nucleotide sequences coding for the shared proteome from Glandora prostrata subsp. lusitanica isolate BPTPS049 and all 12 verified and complete chloroplast genomes belonging to the Boraginaceae family available in GenBank (accession date: 1 June 2022; see supplemental material for additional details). Numbers attached to the branches show the SH-aLRT and the UFBoot2 per cent supports (SH-aLRT/UFBoot2). Ehretia dicksonii (Ehretiaceae), Tiquilia plicata (Ehretiaceae), Salvia rosmarinus (Lamiaceae), and Tilia platyphyllos (Malvaceae) were used as outgroups to the Boraginaceae family (see supplemental material for additional details).
Supplemental Material
Download PDF (2.5 MB)Supplemental Material
Download MS Word (3.1 MB)Data availability statement
The data supporting this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number ON641304. The NCBI BioProject and BioSample are PRJNA848680 and SAMN28118496, respectively. The ENA BioProject and SRA for the generated reads are PRJEB55314 and ERR10047929, respectively.
