Figures & data
Figure 1. Image of S. chienii species. Yukun Wei collected the image in Tangkou Town, Huangshan, Anhui, China (GPS: E114°04′03.15″,N31°49′46.46″). The leaves of S. chienii are long oval with serrated edges. Verticillasters form extended racemes with lavender flowers.

Figure 2. The chloroplast genome map of S. chienii chlobroplast genome. The length of genome is presented in the inner circle. Large single copy (LSC), Single short copy (SSC) and inverted repeat regions (IRA and IRB) are marked. A total of 143 annotated genes are presented in the outer circle, consisting of 91 protein-coding genes, eight rRNA genes, and 37 tRNA genes. Genes were classified by their function in colors. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and Palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. Black: c (complex repeat); Green: p1 (repeat unit size = 1); Yellow: p2 (repeat unit size = 2); Purple: p3 (repeat unit size = 3); Blue: p4 (repeat unit size = 4); Orange: p5 (repeat unit size = 5); Red: p6 (repeat unit size = 6). The small single-copy (SSC), inverted repeat (IRA and IRB), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. Genes are color-coded by their functional classification. The functional classification of the genes is shown in the bottom left corner.
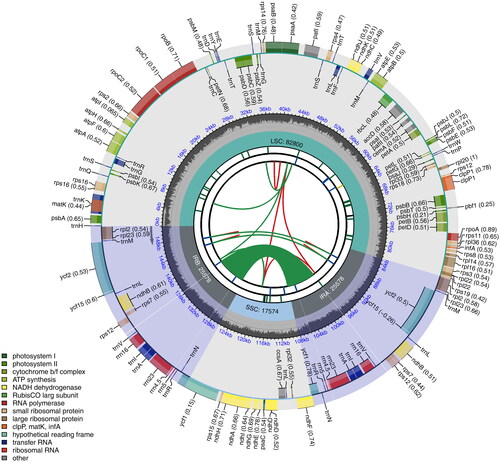
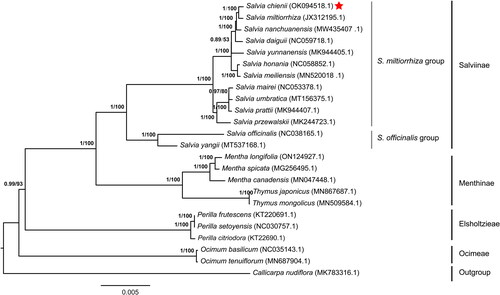
Figure 3. Phylogeny of Lamiaceae based on complete chloroplast genomes, accession numbers were listed behind each taxon. Statistical support values were shown on nodes. The complete chloroplast genomes used for constructing the phylogenetic tree contains S. miltiorrhiza (Hu et al. Citation2020) (JX312195.1), Callicarpa nudiflora (Wang et al. Citation2019) (MK783316.1), Mentha canadensis (Huaizhu et al. Citation2019) (MN047448.1), Mentha longifolia (Zubair Filimban et al. Citation2022)(ON124927.1), Mentha spicata (Wang et al. Citation2017) (MG256495.1), Ocimum basilicum (Rabah et al. Citation2017) (NC035143.1), Ocimum tenuiflorum (Kavya et al. Citation2021) (MN687904.1), Perilla citriodora (Mo et al. Citation2017) (KT22690.1), Perilla frutescens (Shen et al. Citation2016) (KT220691.1), Perilla setoyensis (NC030757.1; Unpublished), Salvia daiguii (Zhou et al. Citation2020) (NC059718.1), Salvia honania (Wang et al. Citation2022) (NC058852.1), Salvia mairei (NC053378.1; Unpublished), Salvia meiliensis (Su et al. Citation2022) (MN520018.1), Salvia nanchuanensis (Su et al. Citation2022) (MW435407.1), S. officinalis (Du et al. Citation2022) (NC038165.1), Salvia prattii (MK944407.1; Unpublished), Salvia przewalskii (Du et al. Citation2019) (MK344723.1), Salvia umbratica (MT156375.1; Unpublished), Salvia yangii (Cao et al. Citation2020) (MT537168.1), Salvia yunnanensis (Tao et al. Citation2019) (MK944405.1), Thymus japonicus (Kim et al. Citation2020) (MN867687.1), Thymus mongolicus (Huaizhu et al. Citation2020) (MN509584.1). The type of sequences used for building the phylogenetic tree are the complete chloroplast genomes downloaded from NCBI (https://www.ncbi.nlm.nih.gov/), and built with IQTree with the best predicted model TVM + F+R3 and 1000 bootstrap replicates. Bayesian posterior probabilities and bootstrap scores were present on the tree nodes. Bayesian posterior probabilitis were culculated by Mrbayes (v3.2.7). S. chienii is close to S. miltiorrhiza.
Supplemental Material
Download JPEG Image (194.7 KB)Supplemental Material
Download JPEG Image (409.6 KB)Supplemental Material
Download JPEG Image (717.7 KB)Data availability statement
The genome sequence data supporting this study’s findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov) under accession no. OK094518. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA763313, SRR15901094, and SAMN21437815, respectively.
