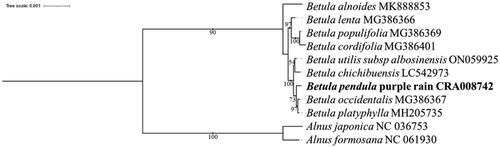
Figures & data
Figure 1. Photo taken at the Forest Genetics and Breeding Base of Northeast Forestry University on July 30, 2022. Eight-year-old B. pendula purple rain.

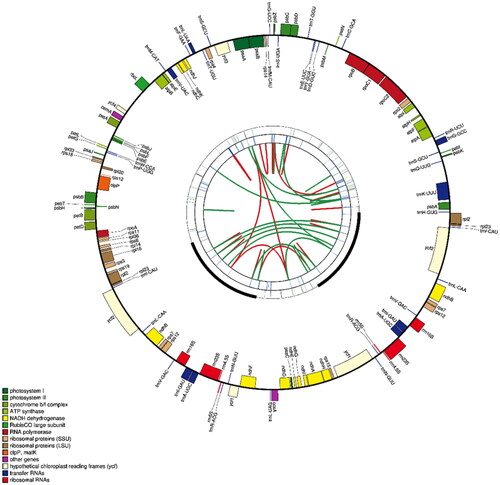
Figure 2. The map contains four rings. From the center going outward, the first circle shows the forward and reverse repeats connected with red and green arcs respectively. The next circle shows the tandem repeats marked with short bars. The third circle shows the microsatellite sequences identified using MISA. The fourth circle is drawn using drawgenemap and shows the gene structure on the plastome. The genes were colored based on their functional categories.
Data availability statement
Data availability statement The genome sequence data that support the findings of this study are openly available in the National Genomics Data Center of NGDC at https://ngdc.cncb.ac.cn/ under the accession no. CRA008742. The associated BioProject, GSA, and Bio-Sample numbers are PRJCA012943, CRA008742, and SAMC976969, respectively.