Figures & data
Figure 1. Gortyna guizhouensis Wu, Yang, and Han, Citation2022 (Lepidoptera, Noctuidae): (A) Larva. The larva is black and orange-yellow; (B) Adult. The ground color of the forewings is golden-red and veins are indistinct; a dark streak is present between the orbicular and reniform spots. Photos by Meng Leng.

Figure 2. Read coverage map across the G. guizhouensis mitochondrial genome. Circlize v0.4.16 was used for plotting.
Figure 3. Location and arrangement of genes on the mitogenome of the G. guizhouensis. A circular mitochondrial genome map was drawn using the Proksee server (https://proksee.ca/).
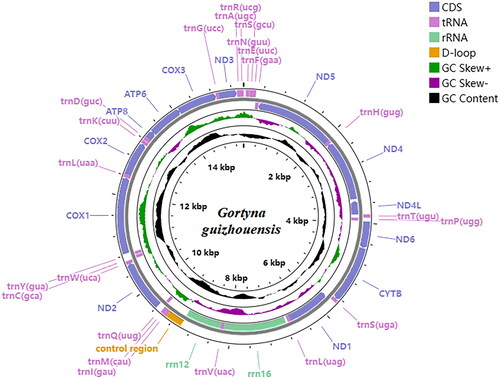
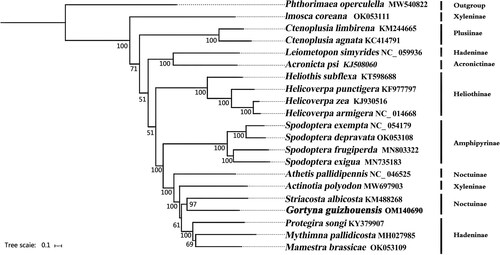
Figure 4. Phylogenetic tree based on 13 mitochondrial protein-coding genes of 21 Lepidoptera species reconstructed by Bayesian inference (BI) method. Numbers on branches are Bootstrap support values (BS). The following sequences were used: MW540822 (Song et al. Citation2021), OK053111 (unpublished), KM244665 (unpublished), KC414791 (Gong et al. Citation2013), NC_059936 (unpublished), KJ508060 (unpublished), KT598688 (De Souza et al. Citation2016), KF977797 (Walsh Citation2016), KJ930516 (Perera et al. Citation2016), NC_014688 (Kômoto et al. Citation2011), NC_054179 (Li et al. Citation2021), OK053108 (unpublished), MN803322 (unpublished), MN735183 (unpublished), NC_046525 (Li et al. Citation2020), MW697903 (unpublished), KM488268 (Coates & Abel Citation2016), KY379907 (Zhao et al. Citation2022), MH027985 (unpublished), OK053109 (unpublished).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nih.gov under the accession no. OM140690. The associated BioProject, SRA and BioSample numbers are PRJNA826860, SRS12616393, and SAMN27585214, respectively.
