Figures & data
Figure 1. A reference image of Euphaea ochracea sequenced in this work, collected from Ayer Hitam Forest Reserve Johor, Malaysia.

Figure 2. Draft mitogenome map of E. ochracea generated in this study. Genes encoded on the heavy strand are transcribed in a clockwise manner, while those encoded on the light strand are transcribed in an anti-clockwise manner. Protein-coding genes are indicated in greyish-blue, transfer RNAs in purple and ribosomal RNAs in green. The innermost circle represents the mitogenome coordinates (Kbp).
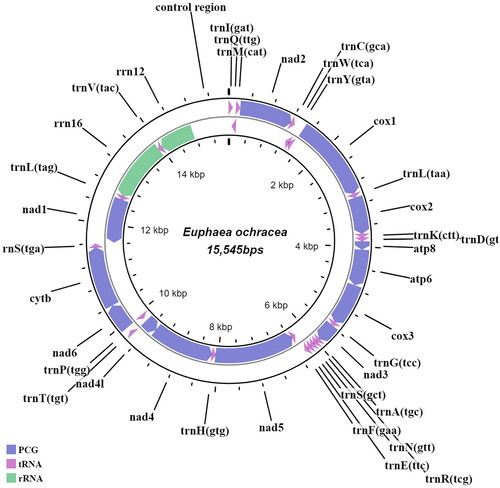
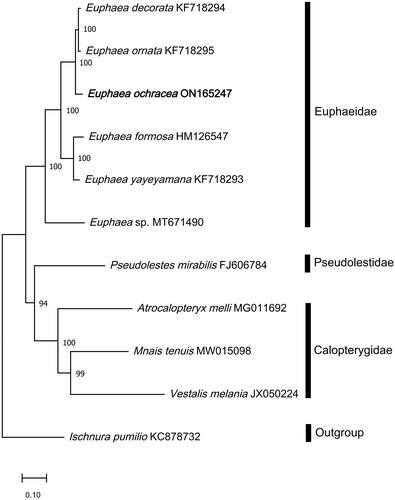
Figure 3. Phylogenetic analysis based on Maximum-Likelihood of 11 Odonata mitogenome sequences, including the newly sequenced E. ochracea, using 13 concatenated protein-coding genes (PCGs). All congeneric mitogenome sequences from Euphaeidae was used in the analyses. Nodal support values indicate the Maximum-Likelihood bootstrap support value (BP). The newly sequenced E. ochracea is highlighted in bold. Mnais tenuis (MW15098) (Wang et al. Citation2021); Vestalis Melania (JX050224) (Chen et al. Citation2015); Atrocalopteryx melli (MG011692) (Xu et al. Citation2018); Euphaea decorata (KF718294), Euphaea ornata (KF718295), Euphaea yayeyamana (KF718293) (Cheng et al. Citation2018); Euphaea sp. (MT671490) (Macher et al. Citation2020); Euphaea ochracea (ON165247) (this study); Euphaea formosa (HM126547) (Lin et al. Citation2010); Pesudolestes mirabilis (FJ606784) (unpublished); Ischnura pumilio (KC878732) (Lorenzo-Carballa et al. Citation2014).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession no. ON165247. The associated BioProject, SRA and BioSample numbers are PRJNA753627, SRR15422665 and SAMN20720553 respectively.
