Figures & data
Figure 1. Photograph of Daphne pseudomezereum var. koreana. (A) Habit. (B) Flowers. Photo credit: Jae-Jin Lee.

Figure 2. Circular map of the complete chloroplast genome of Daphne pseudomezereum var. koreana. The center of the map indicates the name of the species and the length of its chloroplast genome. Going outward, the first circle shows LSC, SSC, IRa, and IRb with their length. The second circle displays the GC ratio depicted as the proportion of the shaded parts of each section. The third circle shows the gene names with the colors based on their functional categories provided in the lower left of the circular map. Genes inside the circle are transcribed in a clockwise direction, and those outside are in a counterclockwise direction.
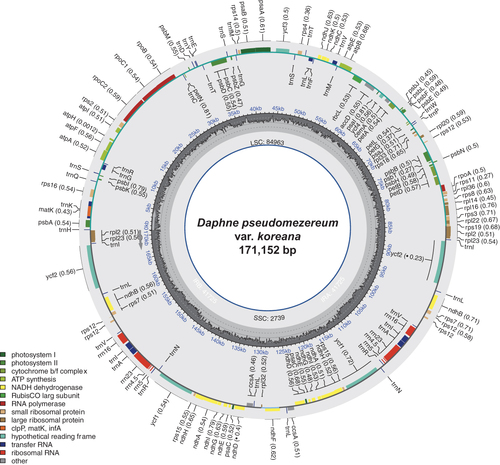
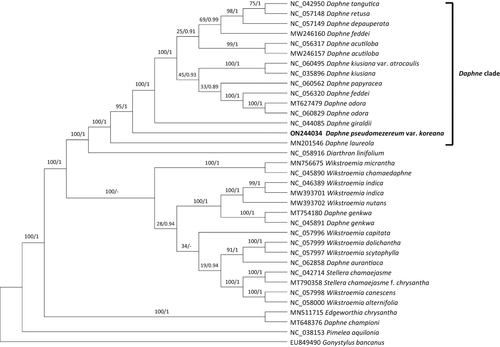
Figure 3. Phylogenetic analyses of 35 whole chloroplast genomes of Daphne and closely related species with Pimelea and Gonystylus as outgroups using the maximum likelihood (ML) and Bayesian inference (BI) methods. The following sequences are used: EU849490 (unpublished), MN201546 (Könyves et al. Citation2019), MN511715 (Qian et al. Citation2021), MN756675 (He et al. Citation2021), MT627479 (Lee et al. Citation2022), MT648376 (Lee et al. Citation2022), MT754180 (Yoo et al. Citation2021), MT790358 (Liang et al. Citation2020), MW246157 (unpublished), MW246160 (unpublished), MW393701 (unpublished), MW393702 (unpublished), NC_035896 (Cho et al. Citation2018), NC_038153 (Foster et al. Citation2018), NC_042714 (Yun et al. Citation2019), NC_042950 (Yan et al. Citation2019), NC_044085 (Yan et al. Citation2019), NC_045890 (Qian et al. Citation2020), NC_045891 (unpublished), NC_046389 (Qian and Zhang Citation2019), NC_056317 (unpublished), NC_056320 (unpublished), NC_057148 (Yan et al. Citation2021), NC_057149 (unpublished), NC_057996 (He et al. Citation2021), NC_057997 (He et al. Citation2021), NC_057998 (He et al. Citation2021), NC_057999 (He et al. Citation2021), NC_058000 (He et al. Citation2021), NC_058916 (Kim et al. Citation2021), NC_060495 (Lee et al. Citation2022), NC_060562 (Lee et al. Citation2022), NC_060829 (unpublished), NC_062858 (unpublished), ON244034 (this study). The phylogenetic tree was drawn based on the ML tree. The numbers above the branches indicate bootstrap support values of ML and the posterior probability from BI.
Supplemental Material
Download MS Power Point (101.4 KB)Data availability statement
The chloroplast genome sequence can be accessed via accession number ON244034 in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA835716, SAMN28108557, and SRR19117239, respectively.
