Figures & data
Figure 1. Species reference image of Spodoptera depravata. The image is referenced from the Bold Systems (Ratnasingham and Hebert Citation2007). Sample ID is SDNU-INS-04111. This figure is attribution noncommercial sharing. http://v3.boldsystems.org/index.php/Taxbrowser_Taxonpage?taxon=Spodoptera+depravata&searchTax=.

Figure 2. Gene map of S. depravata mitochondrial genome. Genes belonging to different functional groups are color-coded. Genes are shown outside and inside the outer circle are transcribed counterclockwise and clockwise, respectively. The grey in the inner circle corresponds to the GC% along the mitochondrion.
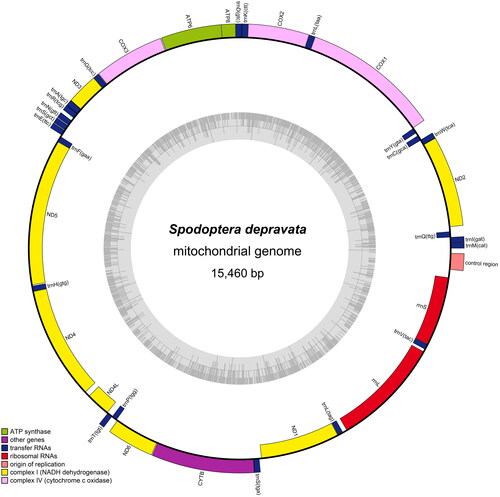
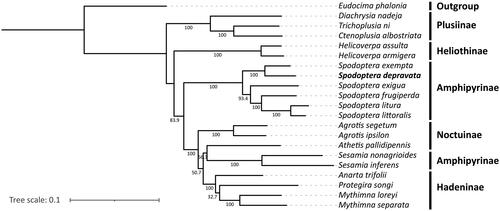
Figure 3. Phylogenetic tree of 21 lepidopteran species based on their mitochondrial genomes. Eudocima phalonia from the family Erebidae was used as the outgroup. The following sequences were used: Eudocima phalonia NC_032382.1 (Seo et al. Citation2019), Diachrysia nadeja MT916722.1 (Gao et al. Citation2021), Trichoplusia ni MK358044.1 (Zhao et al. Citation2022), Ctenoplusia albostriata NC_053742.1 (Xue et al. Citation2019), Helicoverpa assulta NC_014668.1 (Seo et al. Citation2019), Helicoverpa armigera NC_014668.1 (Yin et al. Citation2010), Spodoptera exempta NC_054179.1 (Li, Wang, Xiao, et al. Citation2021), Spodoptera exigua JX316220.1 (Wu et al. Citation2013), Spodoptera frugiperda MN385596.1 (Li, Wang, Xiao, et al. Citation2021), Spodoptera litura NC_022676.1 (Wan et al. Citation2013), Spodoptera littoralis MT816470.1 (Li, Wang, and Zhang Citation2021), Agrotis segetum KC894725.1 (Wu et al. Citation2014), Agrotis ipsilon KF163965 (Zhao et al. Citation2022), Athetis pallidipennis NC_046525.1 (Li, Wang, and Zhang Citation2021), Sesamia nonagrioides CM030612.1 (Muller et al. Citation2021), Sesamia inferens JN039362.1 (Wu et al. Citation2019), Anarta trifolii NC_046049.1 (Li, Wang, and Zhang Citation2021), Protegira songi NC_034938.1 (Guo et al. Citation2017), Mythimna loreyi NC_057500.1 (Nam et al. Citation2020), Mythimna separata KM099034.1 (Li, Wang, and Zhang Citation2021).
Supplemental Material
Download PDF (302.4 KB)Supplemental Material
Download MS Word (673.4 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession no. OK053108. The associated BioProject, BioSample, and SRA numbers are PRJNA809185, SAMN26149759, and SRS12076762 respectively.
