Figures & data
Figure 1. Tree (A) and fruit (B) morphologies of Lonicera caerulea var. edulis.
Note: location, Horticulture Experimental Station (126.73N, 45.74E), Northeast Agricultural University (NEAU), Harbin, China; botanical identification, Pro. Xiuju Wu; photograph, Chenqiao Zhu.

Figure 2. Feature map of Lonicera caerulea var. edulis chloroplast genome.
From the center outward, the first track shows the dispersed repeats, which consist of direct (red arc) and Palindromic (green arc). The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors (Black: complex repeat; Green: 1-unit repeat). The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content is plotted on the fifth track. The annotated genes are shown on the sixth track (codon usage bias is displayed in the parenthesis) and their functional classifications are color-coded in the bottom left corner. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively.
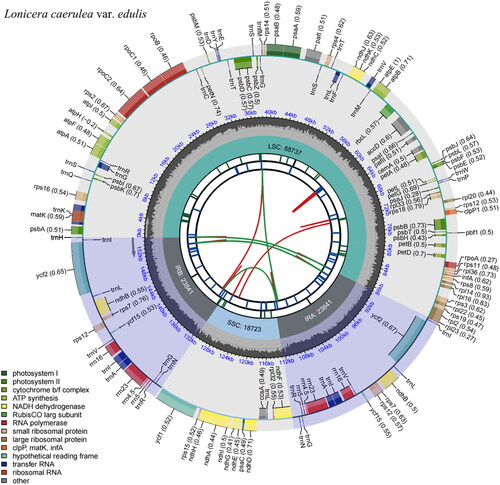
Figure 3. Phylogenetic position of Lonicera caerulea var. edulis inferred from maximum likelihood (ML) based on 33 complete chloroplast genomes.
The subclassification of the Lonicera species examined in this study is according to Rehder (Citation1903, Citation1913), and the representative colors of the subsections are shown on the left diagram. The subfamily classification was labeled according to Wang et al. (Citation2020). The Numbers shown next to the nodes are bootstrap support values based on 1,000 replicates. The following sequences were used: MW795591 (Liu et al. Citation2022b), MZ779026 (Zhang et al. Citation2022), MZ241298 (Jiang et al. Citation2021), NC060471 (Wei et al. Citation2021), MW186760 (Yu et al. Citation2021), MW186761 (Gu et al. Citation2021), MH579750 (Hu et al. Citation2018), MZ901373 (Yang et al. Citation2022), OK393707 (Chen et al. Citation2022), MW296954 (Gu et al. Citation2022), MW340876 (Yuan et al. Citation2021), MN256451 (Jia et al. Citation2020), MW242826, MK176510 (Liu et al. Citation2018), OL457164 (Mo et al. Citation2022), MH681655 (Zhu et al. Citation2019), MK176512 (Liu et al. Citation2018), MH028743 (Kang et al. Citation2018), OP345475 (assembled in the present study), MZ962399 (Wang et al. Citation2022), MK176511 (Liu et al. Citation2018), MG738668 (Fan et al. Citation2018), MG738669 (Fan et al. Citation2018), MH028740 (Kang et al. Citation2018), MG738667 (Fan et al. Citation2018), NC045051 (Wang et al. Citation2020), NC042190, NC029874 (Bai et al. Citation2017), NC045063 (Wang et al. Citation2020), NC045046 (Wang et al. Citation2020), MN524626 (Wang et al. Citation2020), NC033878 (Fan et al. Citation2018), NC032296 (Huang and Cronk Citation2015).
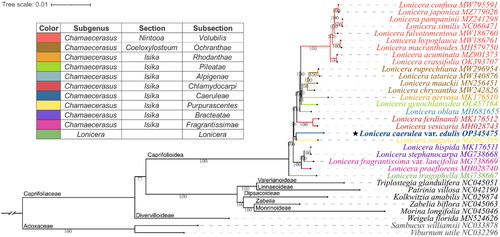
Table 1. Gene annotation summary of Lonicera caerulea var. edulis chloroplast genome.
Supplemental Material
Download MS Word (14.8 MB)Data availability statement
The cp assembly in this study is openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under accession no. OP345475. The associated BioProject, SRA, and BioSample numbers are PRJNA870700, SRR21133392, and SAMN30383140 respectively.
