Figures & data
Figure 1. The reference image of Duoma sheep. Duoma sheep were white or brown coated with the black or brown or mixed color around the eyes and noses, their eyelids and lips are black or brown and occasional pink. Large curved horns are existed in vast majority male Duoma sheep, and female are mainly with small curved or polled. This reference image was taken by Tianzeng Song, the first author listed in this manuscript

Supplemental Material
Download MS Word (34.5 KB)Supplemental Material
Download JPEG Image (381.8 KB)Supplemental Material
Download MS Word (28 KB)Data availability statement
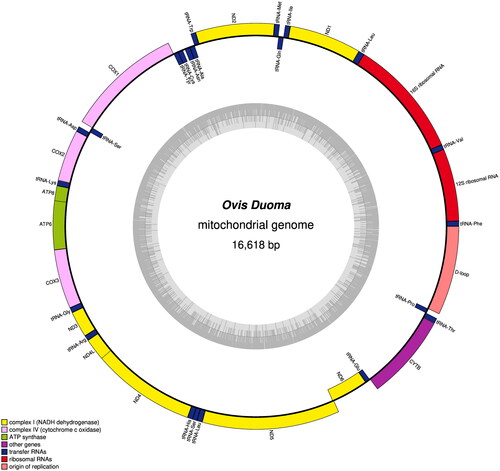
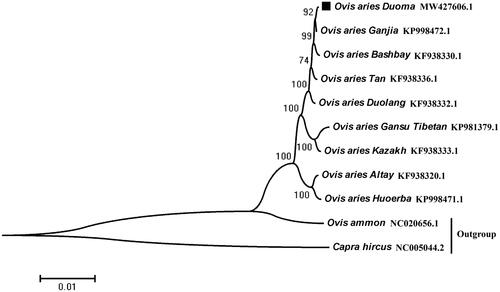
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MW427606.1.