Figures & data
Figure 1. Panorama (A) and detail (B) photos of Bidens pilosa. The photos were shot by Liqiang Wang and the coordinates of the plant was E116.278965, N40.041831. Main identifying traits: Capitulum, margin with tongue-like flower 5–7; tongue elliptic obovate, white, 5–8 mm long; apex obtuse or notched.

Figure 2. Schematic map of overall features of the Bidens pilosa plastome. The map contains six tracks in default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and Palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. Black: c (complex repeat); Green: p1 (repeat unit size = 1); Yellow: p2 (repeat unit size = 2); Purple: p3 (repeat unit size = 3); Blue: p4 (repeat unit size = 4); Orange: p5 (repeat unit size = 5); Red: p6 (repeat unit size = 6). The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
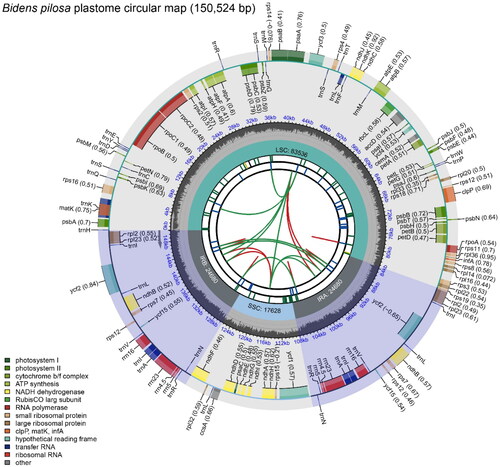
Figure 3. The Maximum-Likelihood phylogeny of Bidens pilosa and its close relatives using 70 common CDS sequences. The bootstrap values based on 1000 replicates were shown on each node in the cladogram tree. The corresponding phylogram tree was shown in the upper right corner (Dots represented Bidens species). The 45 Bidens species Heliantheae alliance were downloaded from GenBank, which were B. pilosa (MZ127828), B. pilosa (MN433104) (Knope et al. Citation2020), B. alba var. radiata (MW551952) (Wu et al. Citation2022), B. pilosa (MW551953) (Wu et al. Citation2022), B. alba var. radiata (MW551955) (Wu et al. Citation2022), B. alba var. radiata (MZ127826), B. bipinnata (MZ127827), B. alba var. radiata (MN433106), B. bipinnata (MW551954), B. bipinnata (MW551957), B. bipinnata (MW551949), B. bipinnata (MW551956), B. biternata (MW691202), B. biternata (MW551958) (Wu et al. Citation2022), B. parviflora (MW551948) (Wu et al. Citation2022), B. hillebrandiana subsp. polycephala (MN433116) (Knope et al. Citation2020), B. forbesii subsp. kahiliensis (MN433094) (Knope et al. Citation2020), B. cosmoides (MN433115) (Knope et al. Citation2020), B. forbesii subsp. forbesii (MN433028) (Knope et al. Citation2020), B. conjuncta (MN433099) (Knope et al. Citation2020), B. campylotheca subsp. pentamera (MN433107) (Knope et al. Citation2020), B. menziesii subsp. menziesii (MN433101) (Knope et al. Citation2020), B. campylotheca subsp. campylotheca (MN433029) (Knope et al. Citation2020), B. amplectens (MN433103) (Knope et al. Citation2020), B. molokaiensis (MN433097) (Knope et al. Citation2020), B. wiebkei (MN433098) (Knope et al. Citation2020), B. valida (MN433092) (Knope et al. Citation2020), B. cervicata (MN433100) (Knope et al. Citation2020), B. hawaiensis (MN433095) (Knope et al. Citation2020), B. micrantha subsp. ctenophylla (MN433114) (Knope et al. Citation2020), B. menziesii subsp. filiformis x B. micrantha subsp. ctenophylla (MN433096) (Knope et al. Citation2020), B. menziesii subsp. filiformis (MN433110) (Knope et al. Citation2020), B. micrantha subsp. kalealaha (MN433102) (Knope et al. Citation2020), B. sandvicensis subsp. confusa (MN433091) (Knope et al. Citation2020), B. sandvicensis subsp. sandvicensis (MN433093) (Knope et al. Citation2020), B. asymmetrica (MN433105) (Knope et al. Citation2020), B. macrocarpa (MN433111) (Knope et al. Citation2020), B. sandvicensis (MN433113) (Knope et al. Citation2020), B. torta (MN433112) (Knope et al. Citation2020), B. mauiensis (MN433090) (Knope et al. Citation2020), B. schimperi (MN433108) (Knope et al. Citation2020), B. pachyloma (MN433109) (Knope et al. Citation2020), B. pilosa (MN729611) (Lin et al. Citation2018), B. tripartita (MW331585) (Wu et al. Citation2022), B. pilosa (MN385242, generated in this study, labeled by bold font and a diamond), B. frondosa (MT178455) (Feifei Li et al. Citation2020). Another two species Lactuca sativa (KT272174) (Jiang et al. Citation2021) and Taraxacum mongolicum (MN164624) (Kim et al. Citation2016) from the Lactuceae served as the outgroups. Previous B. pilosa plastomes deposited in the GenBank were labeled by triangles. All B. pilosa species were located in three clades.
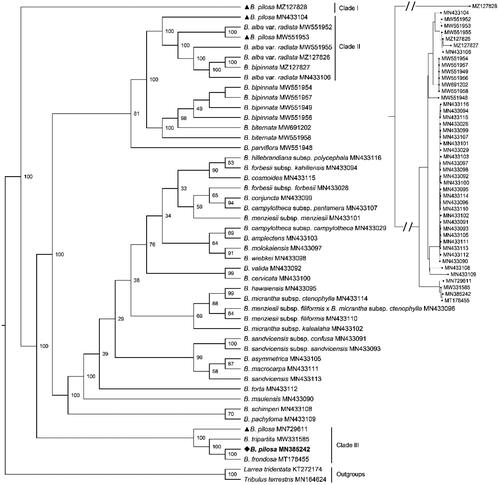
Table 1. Conservative sequences used for designing primers for amplifying potential molecular markers between two B. pilosa plastomes.
Supplemental Material
Download MS Word (248.8 KB)Supplemental Material
Download MS Word (241 KB)Supplemental Material
Download MS Word (20.6 KB)Data availability statement
The plastome sequence has been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with the accession number of MN385242 (https://www.ncbi.nlm.nih.gov/nuccore/MN385242). The associated BioProject, Bio-Sample and SRA numbers are PRJNA543381, SAMN16089020 and SRR12620715 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR12620715).
