Figures & data
Figure 1. The habit and voucher specimen of Amsonia elliptica. (a) Habit of A. elliptica (photo by Bo-Mi Nam, 30 April 2022. Yaksan Island, Wando-gun, South Korea). Amsonia elliptica is a perennial herb and up to 80 cm tall. Leaf blade elliptic, base and apex acuminate; corolla bluish, salverform. (b) The voucher specimen of A. elliptica with a pair of follicles (collected by C. Kim, 4 September 2022, Yaksan Island, Wando-gun, South Korea; herbarium number: HNIBRVP14274 [HIBR]).
![Figure 1. The habit and voucher specimen of Amsonia elliptica. (a) Habit of A. elliptica (photo by Bo-Mi Nam, 30 April 2022. Yaksan Island, Wando-gun, South Korea). Amsonia elliptica is a perennial herb and up to 80 cm tall. Leaf blade elliptic, base and apex acuminate; corolla bluish, salverform. (b) The voucher specimen of A. elliptica with a pair of follicles (collected by C. Kim, 4 September 2022, Yaksan Island, Wando-gun, South Korea; herbarium number: HNIBRVP14274 [HIBR]).](/cms/asset/d362a627-04f4-461a-9bb9-b65cbe626156/tmdn_a_2192834_f0001_c.jpg)
Figure 2. Schematic map of overall features of complete chloroplast genome of Amsonia elliptica. The map contains six circles. From the center outward, the first circle shows the distributed repeats connected with red (forward direction) and green (reverse direction) arcs. The second circle shows tandem repeats marked with short bars. The third circle shows the short tandem repeats of microsatellite sequences as short bars with different colors. The fourth circle displays the sizes of large single-copy (LSC), small single-copy (SSC), and inverted repeat (IRa and IRb) regions. The fifth circle illustrates the distribution of GC contents along the chloroplast genome (dark grey, GC content; light grey, background). The sixth circle displays the genes with colored boxes.
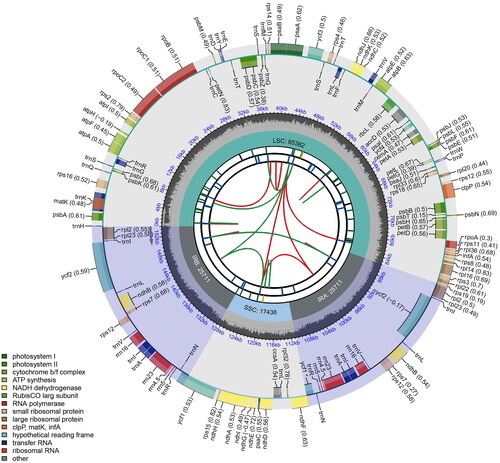
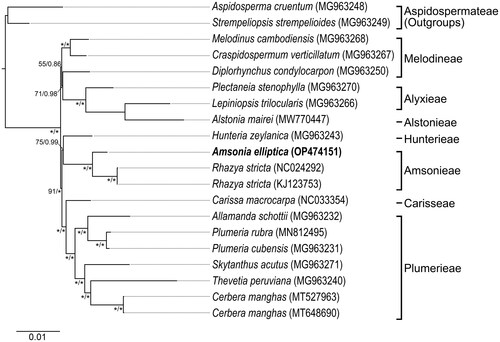
Figure 3. Maximum-likelihood phylogram inferred from 76 cpDNA coding regions of 20 cp genomes from Rauvolfioideae (Apocynaceae). Numbers near nodes indicate support values (maximum likelihood (BS)/Bayesian posterior probability (PP)); an asterisk indicates that a node BS = 100% for maximum-likelihood analysis and PP = 1.00 for Bayesian’s inference analysis. The scale bar shows the number of substitutions per site. Tribes of Rauvolfioideae classification follow Endress et al. (Citation2014). The target species was marked in bold. The following sequences were used: Aspidosperma cruentum MG963248 (Fishbein et al. Citation2018), Strempeliopsis strempelioides MG963249 (Fishbein et al. Citation2018), Melodinus cambodiensis MG963268 (Fishbein et al. Citation2018), Craspidospermum verticillatum MG963267 (Fishbein et al. Citation2018), Diplorhynchus condylocarpon MG963250 (Fishbein et al. Citation2018), Plectaneia stenophylla MG963270 (Fishbein et al. Citation2018), Lepiniopsis trilocularis MG963266 (Fishbein et al. Citation2018), Alstonia mairei MW770447 (unpublished), Hunteria zeylanica MG963243 (Fishbein et al. Citation2018), Amsonia elliptica OP474151 (this study), Rhazya stricta NC024292 (Park et al. Citation2014), KJ123753 (unpublished), Carissa macrocarpa NC033354 (Jo et al. Citation2017), Allamanda schottii MG963232 (Fishbein et al. Citation2018), Plumeria rubra MN812495 (Wang et al. Citation2020), Plumeria cubensis MG963231 (Fishbein et al. Citation2018), Skytanthus acutus MG963271 (Fishbein et al. Citation2018), Thevetia peruviana MG963240 (Fishbein et al. Citation2018), and Cerbera manghas MT527963 (Liao et al. Citation2020), MT648690 (unpublished).
Supplemental Material
Download JPEG Image (1.1 MB)Supplemental Material
Download JPEG Image (511.6 KB)Data availability statement
The genome data that supported the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. OP474151. The associated BioProject, SRA, and Bio-sample numbers are PRJNA885521, SRR21763746, and SAMN31098060, respectively.
