Figures & data
Figure 1. Plant image of Pseudostellaria davidii. This photo was taken by Liqiu Zhang with the author’s approval for use.

Figure 2. Schematic map of overall features of the P. davidii chloroplast genome (Genes drawn outside the outer circle are transcribed clockwise, and those inside are transcribed counter-clockwise. Genes belonging to different functional groups are color-coded. The different colored legends in the bottom left corner indicate genes with different functions. The dark grey inner circle indicates the GC content of the chloroplast genome and the presence of nodes in the LSC, SSC, IR regions).
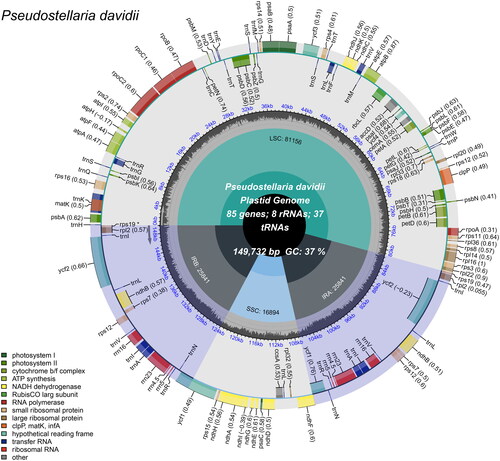
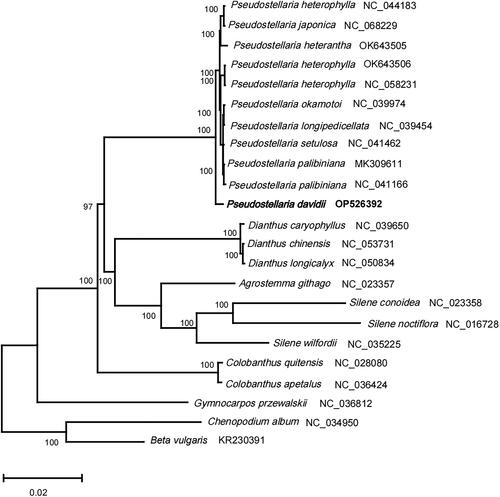
Figure 3. The phylogenetic position for Pseudostellaria davidii according to the ML phylogenetic tree constructed based on 23 chloroplast genomes. The following sequences were used: Pseudostellaria heterophylla NC_044183 (Kim et al. Citation2019b), Pseudostellaria japonica NC_068229, Pseudostellaria heterophylla OK_643505, Pseudostellaria heterophylla OK_643506, Pseudostellaria heterantha NC_058231, Pseudostellaria okamotoi NC_039974, Pseudostellaria longipedicellata NC_039454, Pseudostellaria setulosa NC_041462, Pseudostellaria palibiniana MK_309611 (Kim et al. Citation2019a), Pseudostellaria palibiniana NC_041166, Dianthus caryophyllus NC_039650, Dianthus chinensis NC_053731, Dianthus longicalyx NC_050834, Agrostemma githago NC_023357 and Silene conoidea NC_023358 (Sloan et al. Citation2014), Silene noctiflora NC_016728 (Sloan et al. Citation2012), Silene wilfordii NC_035225, Colobanthus quitensis NC_028080, Colobanthus apetalus NC_036424, Gymnocarpos przewalskii NC_036812, Chenopodium album NC_034950 (Hong et al. Citation2017), Beta vulgaris subsp. vulgaris KR_230391 (Stadermann et al. Citation2015). The sequences used for the tree structure are coding sequences, and the bootstrap support values are shown on the nodes.
Supplemental Material
Download TIFF Image (64.4 MB)Supplemental Material
Download TIFF Image (23.5 MB)Supplemental Material
Download MS Word (379.7 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OP526392. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA903544, SRR22352177, and SAMN31807405, respectively.
