Figures & data
Figure 1. Fruits with winged bracts of Engelhardia hainanensis. This photo was photographed by XYM at Lingyun County in Guangxi.

Figure 2. Schematic circular map of overall features of Engelhardia hainanensis chloroplast genome. Graphic showing features of its plastome was generated using CPGview. The map contains six tracks. From the inner circle, the first track depicts the dispersed repeats connected by red (forward direction) and green (reverse direction) arcs, respectively. The second track shows the long tandem repeats as short blue bars. The third track displays the short tandem repeats or microsatellite sequences as short bars with different colors. The fourth track depicts the sizes of the inverted repeats (IRa and IRb), small single-copy (SSC), and large single-copy (LSC). The fifth track plots the distribution of GC contents along the plastome. The sixth track displays the genes belonging to different functional groups with different colored boxes. The outer and inner genes are transcribed in the clockwise and counterclockwise directions, respectively.
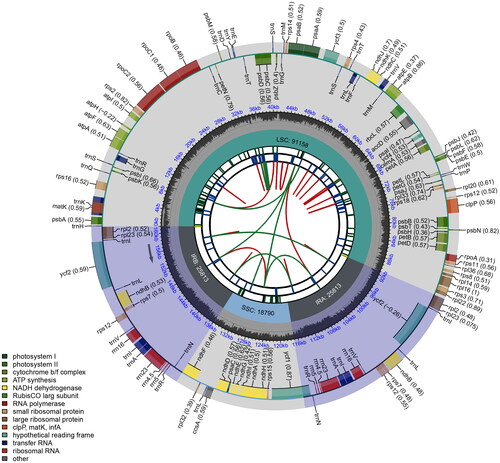
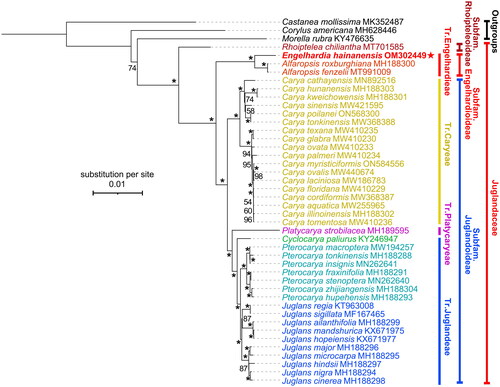
Figure 3. Phylogenetic tree of Juglandaceae inferred from maximum-likelihood (ML) method based on concatenated complete chloroplast genome sequence of 41 species with Corylus americana, Castanea mollissima, and Morella rubra as outgroup. *The clade support value of 100 that was generated from the maximum likelihood method. The red star represents the assembled plastome sequence (i.e., Engelhardia hainanensis OM302449) in this study. GenBank accession numbers of the following sequences were used: Castanea mollissima MK352487 (Zhu et al. Citation2019), Corylus americana MH628446 (Zhao et al. Citation2020), Morella rubra KY476635 (Liu et al. Citation2017), Rhoiptelea chiliantha MT701585 (Jin et al. Citation2020), Alfaropsis roxburghiana MH188300 (Zhou et al. Citation2021), Alfaropsis fenzelii MT991009 (Liu et al. Citation2021), Carya cathayensis MN892516 (Shen et al. Citation2022), Carya hunanensis MH188303 (Zhou et al. Citation2021), Carya kweichowensis MH188301 (Zhou et al. Citation2021), Carya sinensis MW421595 (Xi et al. Citation2022), Carya poilanei ON568300 (Xi et al. Citation2022), Carya tonkinensis MW368388 (Xi et al. Citation2022), Carya texana MW410235 (Xi et al. Citation2022), Carya glabra MW410230 (Xi et al. Citation2022), Carya ovata MW410233 (Xi et al. Citation2022), Carya palmeri MW410234 (Xi et al. Citation2022), Carya myristiciformis ON584556 (unpublished), Carya ovalis MW410234 (Xi et al. Citation2022), Carya laciniosa MW186783 (Zhai et al. Citation2021), Carya floridana MW410229 (Xi et al. Citation2022), Carya cordiformis MW368387 (Xi et al. Citation2022), Carya aquatica MW255965 (Xi et al. Citation2022), Carya illinoinensis MH188302 (Zhou et al. Citation2021), Carya tomentosa MW410236 (Xi et al. Citation2022), Platycarya strobilacea MH189595 (Zhou et al. Citation2021), Cyclocarya paliurus KY246947 (Hu et al. Citation2017), Pterocarya macroptera MW194257 (Yan et al. Citation2021), Pterocarya tonkinensis MH188288 (Zhou et al. Citation2021), Pterocarya insignis MN262641 (Mu et al. Citation2020), Pterocarya fraxinifolia MH188291 (Zhou et al. Citation2021), Pterocarya stenoptera MN262640 (Mu et al. Citation2020), Pterocarya zhijiangensis MH188304 (Zhou et al. Citation2021), Pterocarya hupehensis MH188293 (Zhou et al. Citation2021), Juglans regia KT963008 (Hu et al. Citation2017), Juglans sigillata MF167465 (Dong et al. Citation2017), Juglans ailanthifolia MH188299 (Zhou et al. Citation2021), Juglans mandshurica KX671975 (Hu et al. Citation2017), Juglans hopeiensis KX671977 (Hu et al. Citation2017), Juglans major MH188296 (Zhou et al. Citation2021), Juglans microcarpa MH188295 (Zhou et al. Citation2021), Juglans hindsii MH188297 (Zhou et al. Citation2021), Juglans nigra MH188294 (Zhou et al. Citation2021), Juglans cinerea MH188298 (Zhou et al. Citation2021).
Supplemental Material
Download TIFF Image (391 KB)Supplemental Material
Download TIFF Image (926.4 KB)Supplemental Material
Download TIFF Image (28.6 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OM302449. The associated BioProject, SRA, and BioSample numbers are PRJNA797548, SRP355546 and SAMN25010399, respectively.
