Figures & data
Figure 1. Species reference image of Drechslerella dactyloides. (A) The hypha and conidia (black arrow). (B) The constricting rings (white arrow) for capturing nematodes. Bar = 50 μm. These photographs were taken in this study.
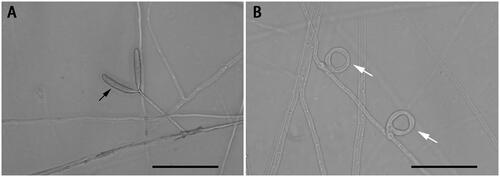
Figure 2. A circle diagram of the assembled mitochondria genome of Drechslerella dactyloides. [1] GC skew. green, GC skew -; red, GC skew+. [2] GC content. red, greater than the genome average GC content; green, less than the genome average GC content. [3] Protein coding genes (red), tRNAs (green) and rRNAs (blue).
![Figure 2. A circle diagram of the assembled mitochondria genome of Drechslerella dactyloides. [1] GC skew. green, GC skew -; red, GC skew+. [2] GC content. red, greater than the genome average GC content; green, less than the genome average GC content. [3] Protein coding genes (red), tRNAs (green) and rRNAs (blue).](/cms/asset/dd0b626d-3e6b-4954-81f1-6afb151cd547/tmdn_a_2197084_f0002_c.jpg)
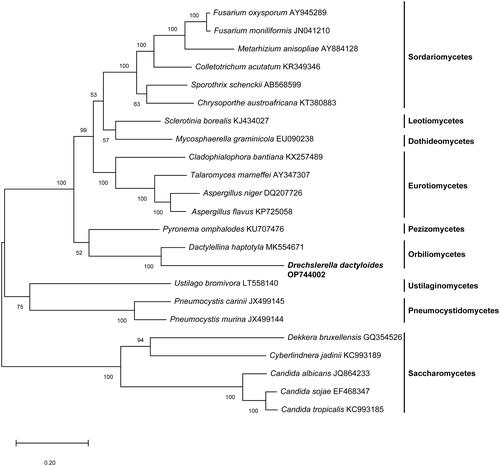
Figure 3. A maximum-likelihood tree of 23 fungal species based on concatenated amino acid sequences of 14 mitochondrial protein amino acid sequences. The 14 mitochondrial protein were: atp6, atp8, atp9, cox1, cox2, cox3, cob, nad1, nad2, nad3, nad4, nad4L, nad5 and nad6. The tree was generated using Mega-X and the best model was “WAG with Freqs. (+F).” Numerical values along branches represent bootstraps based on 1000 randomizations.
Supplemental Material
Download TIFF Image (2.7 MB)Supplemental Material
Download MS Word (572 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under accession no. OP744002. The associated BioProject accession number is PRJNA916072; the Bio-Sample number is SAMN32411356; the SRA accession numbers are SRR23292935 and SRR23292936.
