Figures & data
Figure 1. Species reference image of the Primula vialii. This image is provided by the corresponding author Yuan Huang.

Figure 2. Genomic map of Primula vialii chloroplast genome generated by CPGview (http://www.1kmpg.cn/cpgview/). The species name is shown in the left top corner. The map contains six tracks in default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct and Palindromic repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The small single-copy (SSC), inverted repeat (IRA and IRB), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The base frequency at each site along the genome will be shown between the fourth and fifth tracks. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes belonging to different functional groups are color-coded. Genes drawn inside the circle are transcribed clockwise, and those outside are transcribed counterclockwise.
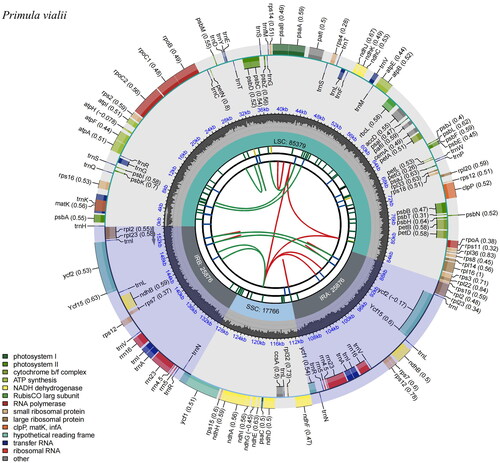
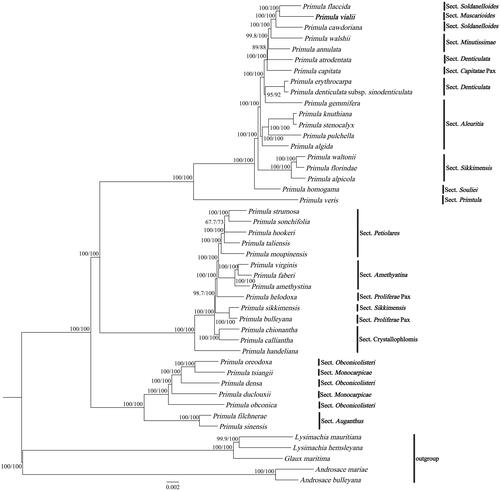
Figure 3. ML phylogenetic tree of Primula vialii and 44 Primulaceae species based on complete chloroplast genomes, the branch supports values were reported as SH-aLRT/UFBoot. The bolded font represents the chloroplast genome of P. vialii in this study. The following sequences were used: Primula flaccida NC_053595 (Wang et al. Citation2021), Primula cawdoriana NC_053586 (Wang et al. Citation2021), Primula walshii NC_053597 (Wang et al. Citation2021), Primula annulata NC_053608 (Wang et al. Citation2021), Primula atrodentata NC_053587 (Wang et al. Citation2021), Primula capitata NC_053589 (Wang et al. Citation2021), Primula erythrocarpa NC_053598 (Wang et al. Citation2021), Primula denticulata subsp. sinodenticulata NC_050247, Primula gemmifera NC_053590 (Wang et al. Citation2021), Primula knuthiana NC_039350 (Ren et al. Citation2018); Primula stenocalyx NC_058249 (Guo et al. Citation2021b), Primula pulchella NC_050246, Primula algida NC_053582 (Wang et al. Citation2021), Primula waltonii NC_058808, Primula florindae NC_053579 (Wang et al. Citation2021), Primula alpicola NC_053588 (Wang et al. Citation2021), Primula homogama NC_054305 (Sun et al. Citation2021), Primula veris NC_031428, Primula strumosa NC_053599 (Wang et al. Citation2021), Primula sonchifolia NC_053594 (Wang et al. Citation2021), Primula hookeri NC_053593 (Wang et al. Citation2021), Primula taliensis NC_053601 (Wang et al. Citation2021), Primula moupinensis NC_050244, Primula virginis NC_053581 (Wang et al. Citation2021), Primula faberi NC_053576 (Wang et al. Citation2021), Primula amethystina NC_053577 (Wang et al. Citation2021), Primula helodoxa NC_046771 (Zhang, Chen, et al. Citation2019), Primula sikkimensis NC_050243, Primula bulleyana NC_046947 (Chen, Zhang, et al. Citation2019), Primula chionantha NC_053583, (Wang et al. Citation2021), Primula calliantha MZ054238 (Yang et al. Citation2021), Primula handeliana NC_039348 (Ren et al. Citation2018), Primula oreodoxa NC_050848, Primula tsiangii NC_046755 (Chen, Yan, et al. Citation2019), Primula densa NC_058262 (Zhong et al. Citation2019), Primula duclouxii NC_058263 (Zhong et al. Citation2019), Primula obconica NC_046415 (Zhang, Yuan, et al. Citation2019), Primula filchnerae NC_051972 (Lu et al. Citation2020), Primula sinensis NC_030609 (Liu et al. Citation2016), Lysimachia mauritiana NC_060700 (Lee et al. Citation2022), Lysimachia hemsleyana NC_052863 (Ying et al. Citation2019), Glaux maritima NC_059901 (Liu et al. Citation2021), Androsace mariae NC_051991 (Guo et al. Citation2021a), Androsace bulleyana (NC_034641).
Supplemental Material
Download JPEG Image (357.4 KB)Supplemental Material
Download PNG Image (24.8 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov/] under accession no. ON584545. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA841839, SRR19391427, and SAMN28626130, respectively.
