Figures & data
Figure 1. Morphology features of C. nitidissima. (a) Individual of C. nitidissima; (b) Mature leaves; (c) Buds and flowers. The photos of C. nitidissima were taken at the nursery of Yulin normal university, Yulin, Guangxi, China.

Figure 2. The gene map of the complete mitochondrial genome of C. nitidissima. Genes indicated in the inner circle are transcribed clockwise, and those indicated in the outer circle are transcribed counterclockwise. Different functional groups of genes are color coded. The darker gray corresponds to DNA G + C content, while the lighter gray corresponds to A + T content.
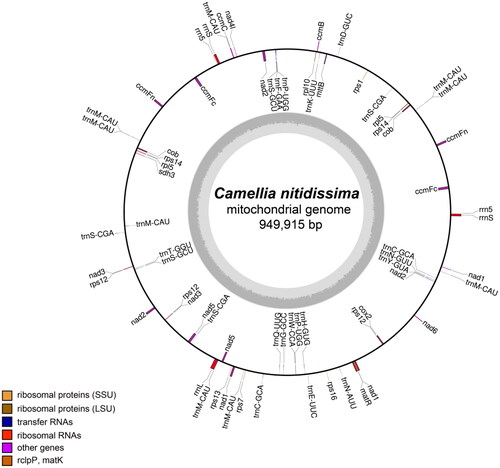
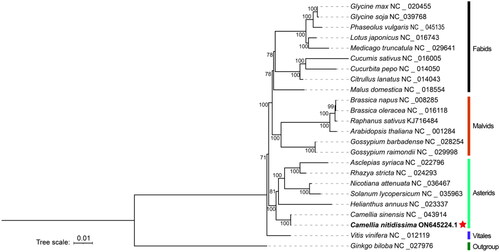
Figure 3. Used the Ginkgo biloba (NC_027976) as outgroup, the phylogenetic tree of 24 plants was constructed with maximum-likelihood method by PhyloSuite. The numbers above the branches were the bootstrap value. The 23 conserved protein-coding genes (atp1, atp4, atp6, atp8, atp9, ccmB, ccmC, cox1, cox3, cytb, matR, mttB, nad1, nad2, nad3, nad4, nad5, nad6, nad7, rpl2, rps10, rps19, rps3) in the mitochondrial genomes were used to construct the phylogenetic.
Supplemental Material
Download MS Word (31.4 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. ON645224.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA899601, SRR22240446, SRR22240447, and SAMN31665678, respectively.
