Figures & data
Figure 1. Eranthis byunsanensis of Wanju, Republic of Korea. (A) Typical radical leaves with blades divided into several palmate segments. (B) White, compound floral structure with broad overlapping sepals and funnel-shaped petals. Scale bars represent 1 cm. Photographs were captured by Joon Moh Park using a digital camera.

Figure 2. Chloroplast genome map of E. byunsanensis. A circular and complete cp genome map was generated by CPGview. Large single-copy, small single-copy, and inverted repeat are represented as LSC, SSC, and IR (IRA and IRB) on the fourth track, respectively. GC content is represented on the fifth track in dark gray. The genes are shown on the sixth track. Genes located on the inner and outer of circle are transcribed clockwise and anticlockwise, respectively. The functional classification of the genes is presented in the bottom left corner.
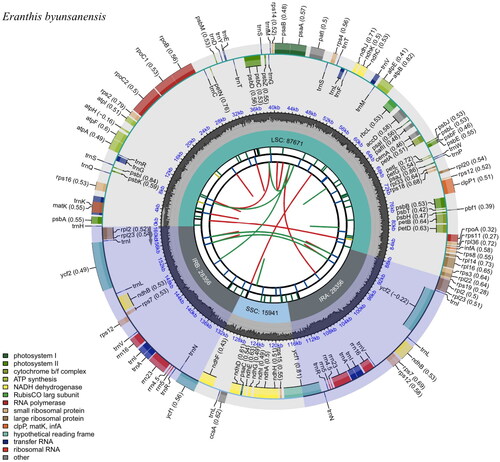
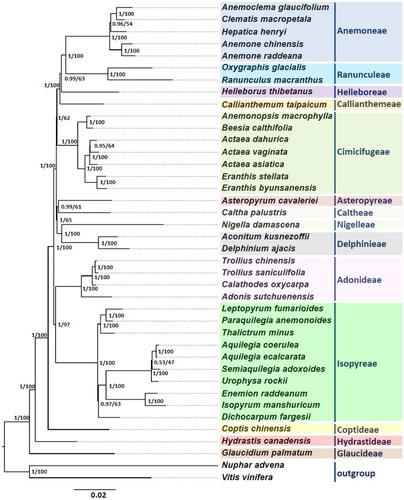
Figure 3. Phylogenetic relationships among 38 Ranunculaceae species based on 77 shared PCGs of the complete cp genome. The sequences used for tree construction are as follows: Anemoclema glaucifolium (MK569471; Zhai et al. Citation2019), Anemone chinensis (MK569491; Zhai et al. Citation2019), Anemone raddeana (MK569472; Zhai et al. Citation2019), Clematis macropetala (MK569482; Zhai et al. Citation2019), Hepatica henryi (MK569494; Zhai et al. Citation2019), Oxygraphis glacialis (MK569489; Zhai et al. Citation2019), Ranunculus macranthus (NC_008796; Raubeson et al. Citation2007), Helleborus thibetanus (MK569493; Zhai et al. Citation2019), Callianthemum taipaicum (MK569479; Zhai et al. Citation2019), Actaea asiatica (MK569469; Zhai et al. Citation2019), Actaea dahurica (MK569481; Zhai et al. Citation2019), Actaea vaginata (MK569499; Zhai et al. Citation2019), Anemonopsis macrophylla (MK569473; Zhai et al. Citation2019), Beesia calthifolia (MK569477), Eranthis stellate (MK569487; Zhai et al. Citation2019), Eranthis byunsanensis (ON564441; this study), Caltha palustris (MK569480; Zhai et al. Citation2019), Asteropyrum cavaleriei (MK569476; Zhai et al. Citation2019), Nigella damascene (MK569488; Zhai et al. Citation2019), Aconitum kusnezoffii (MK569468; Zhai et al. Citation2019), Delphinium ajacis (MK569484; Zhai et al. Citation2019), Adonis sutchuenensis (MK569470; Zhai et al. Citation2019), Calathodes oxycarpa (MK569478; Zhai et al. Citation2019), Trollius saniculifolia (NC_012615; Kim et al. Citation2009), Trollius chinensis (MK569501; Zhai et al. Citation2019), Aquilegia coerulea (MK569474; Zhai et al. Citation2019), Aquilegia ecalcarata (MK569475; Zhai et al. Citation2019), Dichocarpum fargesii (MK569485; Zhai et al. Citation2019), Enemion raddeanum (MK569486; Zhai et al. Citation2019), Isopyrum manshuricum (MK569496; Zhai et al. Citation2019), Leptopyrum fumarioides (MK569497; Zhai et al. Citation2019), Paraquilegia anemonoides (MK569490; Zhai et al. Citation2019), Semiaquilegia adoxoides (MK569498; Zhai et al. Citation2019), Thalictrum minus (MK569500; Zhai et al. Citation2019), Urophysa rockii (MK569502; Zhai et al. Citation2019), Coptis chinensis (MK569483; Zhai et al. Citation2019), Hydrastis Canadensis (MK569495; Zhai et al. Citation2019), Glaucidium palmatum (MK569492; Zhai et al. Citation2019). Nuphar advena (NC_008788; Raubeson et al. Citation2007), and Vitis vinifera (NC_007957; Jansen et al. Citation2006) were used as outgroups. The phylogenetic tree was constructed using BI and ML methods, and the trees showed identical topologies. The numbers on the nodes indicate the BI posterior probability and ML bootstrap value (%), respectively. The scale bar represents the number of substitutions per site.
Supplemental Material
Download MS Word (843.5 KB)Data availability statement
The genome sequence data supporting the findings of this study are openly available as accession no. ON564441 at GenBank in NCBI (https://www.ncbi.nlm.nih.gov). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA835661, SRR19213060, and SAMN28106645, respectively.
