Figures & data
Figure 1. Abies ernestii var. salouenensis. (A) Plant; (B) mature cones; (C) pollen cones; (D) leaves; (E) vegetative buds. M. These images are from Christian (Citation2021) and photography by Yi-Zhen Shao.

Figure 2. Schematic map of overall features of the chloroplast genome of Abies ernestii var. salouenensis. The circular map of the chloroplast genome was generated using CPGview (Liu et al. Citation2023). Genes shown outside the circle are transcribed clockwise, and genes inside are transcribed counter-clockwise. Genes belonging to different functional groups are color-coded. The darker gray in the inner corresponds to the GC content, and the lighter gray to the AT content.
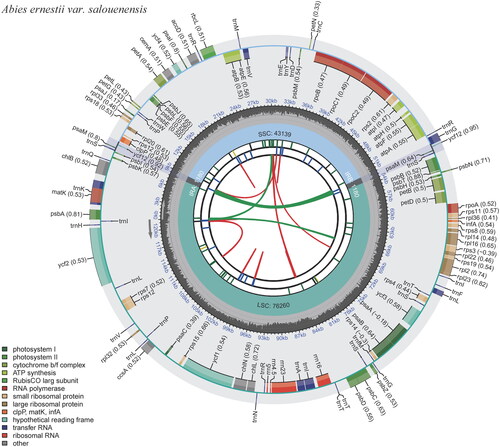
Table 1. List of genes encoded in Abies ernestii var. salouenensis chloroplast genomes.
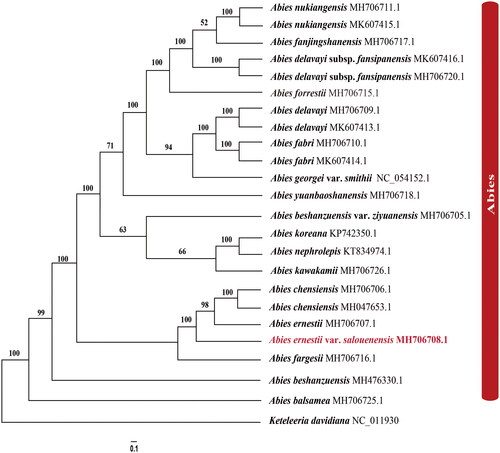
Figure 3. The best Maximum likelihood (ML) phylogram inferred from 23 chloroplast genomes in Abies, with Keteleeria davidiana as an outgroup (bootstrap values are indicated on the branches). The following sequences were used: A. nukiangensis Cheng et L. K. Fu MH706711 and MK607415 (Shao et al. Citation2020), A. fanjingshanensis W. L. Huang, Y. L. Tu and S. Z. Fang MH706717 (Guo et al. Citation2019), A. delavayi subsp. fansipanensis (Q.P.Xiang, L.K.Fu and Nan Li) Rushforth MH706720 and MK607416 (Shao et al. Citation2020), A. forrestii C. C. Rogers MH706715 (Dong et al. Citation2021), A. delavayi Franch. MH706709 and MK607413 (Shao et al. Citation2020), A. fabri (Mast.) Craib MH706710 and MK607414 (Shao et al. Citation2020), A. georgei var. smithii (Viguie et Gaussen) Cheng et L NC_054152 (Li et al. Citation2021), A. yuanbaoshanensis Y. J. Lu & L. K. Fu MH706718 (Zhang et al. Citation2020), A. beshanzuensis var. ziyuanensis (L. K. Fu & S. L. Mo) L. K. Fu & Nan Li MH706705 (Fu et al. Citation2019), A. koreana E. H. Wilson KP742350 (Yi et al. Citation2015), A. nephrolepis (Trautv.) Maxim. KT834974 (Yi et al. Citation2016), A. kawakamii (Hayata) T. Ito MH706726 (Shao et al. Citation2019), A. chensiensis MH706706 and MH047653 (Liu et al. Citation2018; Su et al. Citation2019), A. ernestii MH706707 (Shao et al. Citation2022), A. fargesii Franch. MH706716 (Guo et al. Citation2019), A. beshanzuensis M. H. Wu MH 476330 (Shao et al. Citation2018), A. balsamea (L.) Mill. MH 706725 (Wu et al. Citation2019), and Keteleeria davidiana NC_011930 (www.ncbi.nlm.nih.gov/).
Supplemental Material
Download TIFF Image (6.6 MB)Supplemental Material
Download TIFF Image (5.5 MB)Supplemental Material
Download TIFF Image (6 MB)Supplemental Material
Download TIFF Image (15.9 MB)Supplemental Material
Download MS Word (4.2 MB)Data availability statement
The genomic sequence data supporting the findings of this work are accessible through GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) with the accession number MH706708. The related BioProject, SRA, and Bio-Sample identifiers are PRJNA790665, SRP351584, and SAMN24219952.
