Figures & data
Figure 1. Morphology and habitat map of Corethrodendron multijugum. (a–c) From Yueliang Bay Park in Guide County, Hainan Tibetan Autonomous Prefecture, Qinghai Province, China (101°43′95″ E, 36°04′61″ N). Photograph by Ying Liu.

Figure 2. The plastome genome map of Corethrodendron multijugum under this study. From the inner circle, the first circle depicts distributed repeats connected by red (forward direction) and green (reverse direction) arcs, respectively. The following circle displays tandem repeats denoted by short blue bars. The sequences of microsatellites are depicted as short green bars. The fourth circle displays the sizes of LCS, SSC, and IR (this plastid lacks annotation of these three regions). The fifth circle illustrates the distribution of GC contents along the plastome (dark grey: GC contents, light grey: background). The sixth circle displays the genes with colored boxes. The outer and inner colored boxes present transcribed clockwise and counterclockwise genes, respectively.
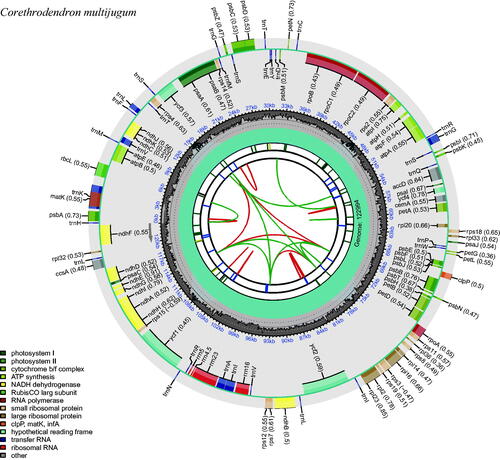
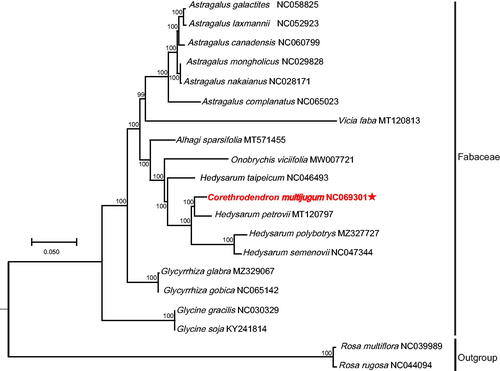
Figure 3. Chloroplast phylogeny of 18 Fabaceae species based on the complete chloroplast genome sequences. The asterisk represents the assembled plastome sequence in this study. The clades of species are represented with black lines. The following sequences of each species were used: Astragalus canadensis NC060799 (unpublished), A. complanatus NC065023 (unpublished), A. galactites NC058825 (Ding et al. Citation2021), A. laxmannii NC052923 (Liu et al. Citation2020), A. mongholicus NC029828 (Lei et al. Citation2016), A. nakaianus NC028171 (Choi et al. Citation2016), Glycine gracilis NC030329 (Gao and Gao Citation2017), G. soja KY241814 (unpublished), Glycyrrhiza glabra MZ329067 (unpublished), G. gobica NC065142 (unpublished), Corethrodendron multijugum NC069301 (this study), Hedysarum petrovii MT120797 (unpublished), H. polybotrys MZ327727 (Cao et al. Citation2021), H. semenovii NC047344 (Zhang et al. Citation2020), H. taipeicum NC046493 (She et al. Citation2019), Vicia faba MT120813 (unpublished), Onobrychis viciifolia MW007721 (Fu et al. Citation2021), Alhagi sparsifolia MT571455 (Wang et al. Citation2020), Rosa multiflora NC039989, and R. rugosa NC044094 (Kim et al. Citation2019). Unpublished in the legend indicates that the citations have not been published.
Supplemental Material
Download PDF (305.7 KB)Supplemental Material
Download PDF (1.4 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank database of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. NC069301 (https://www.ncbi.nlm.nih.gov/nuccore/NC_069301.1). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA896184 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA896184), SRA: SRR22200104 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR22200104), and SAMN31536048 (https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN31536048), respectively.
