Figures & data
Figure 1. The morphology of Limnophila sessiliflora. (A) Transverse rhizome with erected branches, the aerial leaves are simple pinnate and whorled, (B) fruit, and (C) the flowers are pine, with beard in the corolla tube. All were photographed by Hongjin Dong in Xishui, the same location as the specimens collected. A and B were taken in 19 January 2021, and C was taken in 17 June 2021.

Figure 2. The chloroplast genome map of L. sessiliflora, which was generated using CPGview (Liu et al. Citation2023). The center of the map notes the name and the length of the species, also includes the summary data of the chloroplast genome. Going outward, the first circle shows LSC, SSC, IRa, and IRb with their length. The second circle shows the GC ratio depicted as the proportion of the shaded parts of each section. The third circle displays the gene names with the colors based on their functional categories provided at the left of the circular map. Genes outside the circle are transcribed in an anticlockwise direction, and those inside are in a reverse direction.
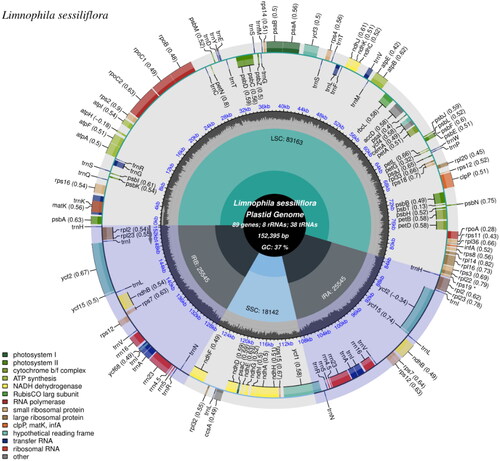
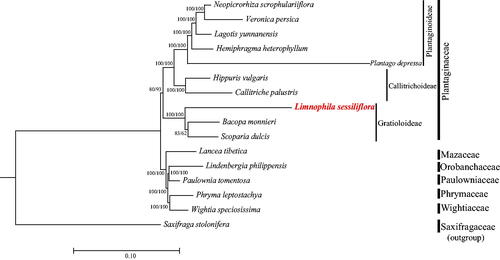
Figure 3. Maximum-likelihood phylogenetic tree including Limnophila sessiliflora based on complete chloroplast genomes. The number at each node indicates the bootstrap support value generated by RAxML/IQ-tree. The following sequences are used: Neopicrorhiza scrophulariiflora MK986819 (Zhang et al. 2019), Veronica persica KT724052 (Choi et al. 2016), Lagotis yunnanensis MN752238 (Cheng et al. 2021), Hemiphragma heterophyllum MN383192 (Wu and Zhang 2019), Plantago depressa MK144833 (Kwon et al. 2019), Hippuris vulgaris MT942637 (Liu et al. 2021), Callitriche palustris MW774642 (Yu and Dong Citation2021), Bacopa monnieri MN736955 (Liang et al. 2020), Scoparia dulcis MZ242235 (Li et al. Citation2022), Lancea tibetica MF593117 (Chi et al. 2018), Lindenbergia philippensis HG530133 (Wicke et al. 2013), Paulownia tomentosa KP718624 (Yi and Kim 2016), Phryma leptostachya MK381317 (Xia et al. 2019), Wightia speciosissima MK381318 (Xia et al. 2019), and Saxifraga stolonifera MN496079 (Dong et al. 2018). The GenBank accession numbers for each species and the citation sources for those published sequences are provided in Table S1.
Supplemental Material
Download MS Word (24.8 KB)Supplemental Material
Download MS Word (616.4 KB)Supplemental Material
Download JPEG Image (486.1 KB)Supplemental Material
Download JPEG Image (1.3 MB)Supplemental Material
Download JPEG Image (1.2 MB)Data availability statement
The data that support the findings of this study are available in the NCBI GenBank database at https://www.ncbi.nlm.nih.gov under accession number ON000200. The assembled individual was linked with BioSample number SAMN26570373 and Project ID PRJNA814836. Raw sequencing reads used in this study were deposited in the GenBank Sequence Read Archive with no. SRR18313725.
