Figures & data
Figure 1. The morphological characteristics of the Cenchrus alopecuroides. (a) the characteristics of whole plant during flowering; (b) the flowering period of C. alopecuroides; (c) the fruiting period of C. alopecuroides.

Figure 2. The chloroplast genome coverage depth and map of Cenchrus alopecuroides. (a) The mapped read depth of Cenchrus alopecuroides chloroplast genome. (b) The chloroplast genome map of Cenchrus alopecuroides. The map contains six circles. From the center to outward, the first circle shows the distributed repeats connected with red (forward) and green (reverse) arcs. The second circle shows the tandem repeats as short bars. The third circle shows the microsatellite sequences marked with short bars. The fourth circle shows the size of the LSC, SSC, and IRs. The fifth circle shows the GC contents along the plastome. The sixth circle shows the genes having different colors based on their functional groups. Genes shown outside the circle are transcribed clockwise, and those inside counter-clockwise.
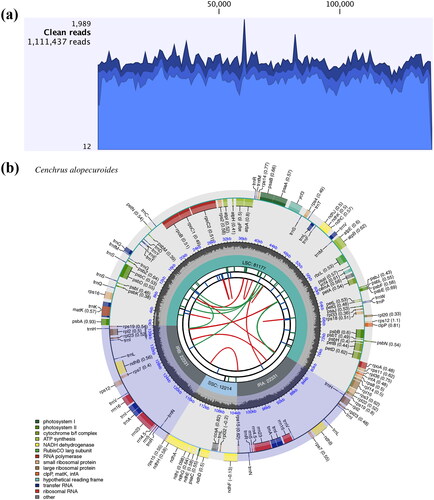
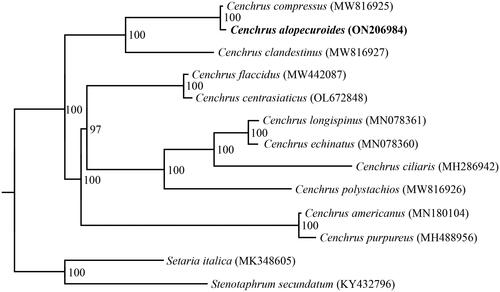
Figure 3. Phylogenetic tree inferred by the maximum likelihood (ML) method based on complete chloroplast genomes of 11 Cenchrus species, with Stenotaphrum secundatum (KY432796) and Setaria italica (MK348605) as outgroups. Numbers near the nodes represent ML bootstrap values. The following sequences were used: MW816925–MW816927 (Xu et al. Citation2021), MW442087 (Liu, W., Direct submission), OL672848 (Zhang, L., Direct submission), MN078360–MN078361 (Hyun et al. Citation2019), MH286942 (Bhatt and Thaker Citation2018), MN18014 (Xu et al. Citation2019), MN488956 (Bhatt and Thaker Citation2018), MK348605 (Liu et al. Citation2019), KY432796 (Gallaher et al. Unpublished).
Supplemental Material
Download TIFF Image (321.6 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. ON206984. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA871033, SRR21166642, and SAMN30399920, respectively.
