Figures & data
Figure 1. The reference image of L. litseifolius. Trees to 20 m tall. Leaf blade elliptic, obovate-elliptic, ovate, or rarely narrowly elliptic, 8–18 × 3–8 cm, papery to sub leathery, margin entire, apex acuminate to acute. Its tender leaves are sweet, commonly known as sweet tea. Female and androgynous inflorescences are usually 2–6 congested at apex of branches, spiciform, to 35 cm; cupules in clusters of 3–5. This image was taken by Xiaoyan Qiu in Liangyaping Town, China (27°48′7.65″N, 110°35′6.73″E).

Figure 2. Maximum-likelihood phylogenetic tree for L.litseifolius was constructed based on 25 complete chloroplast genomes using Corylus fargesii and Corylus heterophylly as outgroups. The following sequence was used: Lithocarpus hancei MW375417.1 (Ma et al. Citation2021), Lithocarpus polystachyus MK914534.1 (Li et al. Citation2019), Lithocarpus konishii ON422319.1 (unpublished), Lithocarpus litseifollus OM048987.1 (unpublished), Lithocarpus balansae KP299291.1 (Li et al. Citation2018), Lithocarpus longinux OK181903.1 (Wu et al. Citation2022), Lithocarpus dealbatus MZ322408.1 (Shelke et al. Citation2022), Castanopsis carlesii MK745999.1 (Liu et al. Citation2019), Castanopsis mekongensis MW043480.1 (unpublished), Castanopsis sieboldii MZ028444.1 (Park et al. Citation2021), Castanopsis sclerophylla MK387847.1 (Shelke et al. Citation2022), Castanea sativa MW044606.1 (unpublished), Castanea henryi MH998384.1 (Gao et al. Citation2019), Castanea mollissima MW322901.1 (Zhang et al. Citation2021), Castanea crenata MW044605.1 (Jeong et al. Citation2019), Quercus bawanglingensis MK449426.1 (Ma et al. Citation2021), Quercus haronii KT963087.1 (Ma et al. Citation2021), Quercus variabilis KU240009.1 (Li et al. Citation2018), Trigonobalanus doichangensis KF990556.1 (Park and Oh Citation2020), Fagus sylvatica MW537046.1 (Mishra et al. Citation2021), Fagus engleriana MT762293.1 (unpublished), Fagus japonica MT762296.1 (unpublished), Corylus fargesii KX822767.2 (Hu et al. Citation2017), and Corylus heterophylla KX822769.2 (Ma et al. Citation2021).
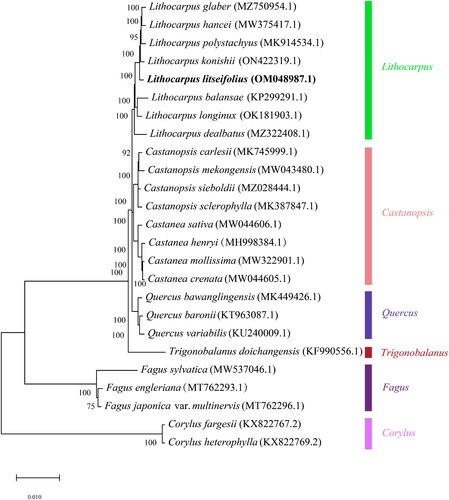
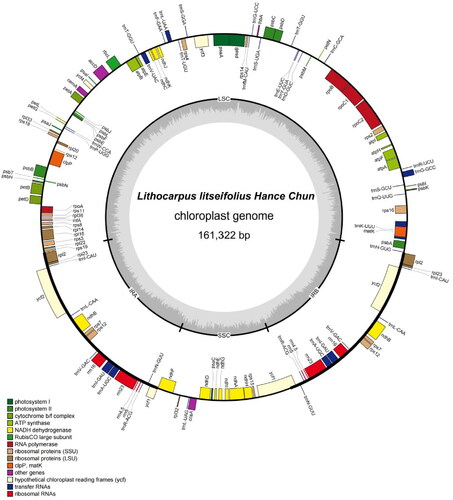
Figure 3. The circular map of L. litseifolius chloroplast genome. Genes with different functions are shown in different colors. Genes shown on the outside and inside of the circle are transcribed clockwise and counterclockwise, respectively. The grey circle inside represents the GC content. The SSC and LSC regions are separated by IRs (IRA and IRB).
Supplemental Material
Download MS Word (6.7 MB)Supplemental Material
Download PNG Image (306.7 KB)Supplemental Material
Download PNG Image (501.1 KB)Supplemental Material
Download PNG Image (5.8 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number OM048987.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA792474, SRR17333522, and SAMN24425764, respectively.
