Figures & data
Figure 1. The specimen and morphology of C. soulattri. (A) The specimen of C. soulattri (accession number: 14288); (B) The fruit and (C) the inflorescence and leaves of C. soulattri. (Photo credits: Jessica Rey).

Figure 2. The chloroplast genome (plastome) map of C. soulattri. The plastome map is divided into six circles with different representations. Starting from the inner circle, the first circle shows the dispersed repeats connected by red (forward repeat) and green (palindromic repeat) arcs. The second circle shows long tandem repeats in short blue bars. The third circle presents simple sequence repeats (SSRs) that are color coded based on repeat unit size (RUS) (Black, complex repeat; Green, RUS = 1; Yellow, RUS = 2; and Blue, RUS = 4). The fourth circle displays the four regions of the plastome (LSC, SSC, IRa, and IRb) with their respective sizes. The fifth circle provides the GC content along the genome. The sixth circle provides the genes with their codon usage bias in parentheses and are color-coded, indicating their respective functional group. The functional groups can be found at the bottom left corner. Genes that are found in the inner and outer circle are transcribed clockwise and counterclockwise, respectively.
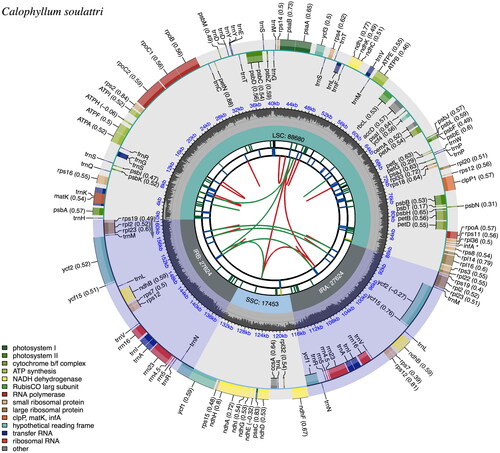
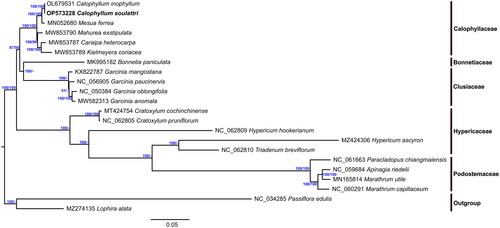
Figure 3. Maximum likelihood (ML) phylogenetic tree and Bayesian inference tree of C. soulattri and other Clusioid species from the order Malpighiales based on the whole chloroplast genome sequence with L. alata and P. edulis as outgroup. Values on the right represents bootstrap support from Maximum Likelihood analysis while on the left represents Bayesian posterior probabilites. The species shown in bold font is newly sequenced in this study. The following sequences were used: C. inophyllum OL679531, M. ferrea MN052680 (Wang et al. Citation2019), Bonnetia paniculata MK995182 (Jin et al. Citation2020), Garcinia mangostana KX822787 (Jo et al. Citation2017), G. paucinervis MT501656 (Wang et al. Citation2021), G. oblongifolia MT726019 (Ma et al. Citation2020), G. anomala MW582313 (Yue and Shi Citation2021), Caraipa exstipulata MW853790 (Trad et al. Citation2021), Kielmeyera coriacea MW853789 (Trad et al. Citation2021), C. heterocarpa MW853787 (Trad et al. Citation2021), Apinagia riedelii MN165812 (Bedoya et al. Citation2019), Marathrum utile MN165814 (Bedoya et al. Citation2020), M. capillaceum MN165813 (Bedoya et al. Citation2020), Paracladopus chiangmaiensis MZ645928 (Wu et al. Citation2022), Hypericum ascyron MZ424306 (Claude et al. Citation2022), Triadenum breviflorum MZ714016 (Sudmoon et al. Citation2022), H. hookerianum MZ714015 (Sudmoon et al. Citation2022), Cratoxylum pruniflorum MZ703416 (Sudmoon et al. Citation2022), C. cochinchinense MT424754 (Huang et al. Citation2019), Lophira alata MZ274135 (Mascarello et al. Citation2021), and NC_034285 Passiflora edulis (Cauz-Santos et al. Citation2017). NCBI accession numbers are given for Genbank sequences.
Supplemental Material
Download PDF (258.4 KB)Supplemental Material
Download JPEG Image (925.8 KB)Supplemental Material
Download JPEG Image (905.3 KB)Supplemental Material
Download PNG Image (951.9 KB)Supplemental Material
Download JPEG Image (553.1 KB)Data availability statement
The chloroplast genome sequence data that support the findings of this study are openly available in GenBank of NCBI database at (https://www.ncbi.nlm.nih.gov/) under the accession no. OP573228. The associated BioProject, SRA, and Biosample numbers of C. soulattri are PRJNA891016, SRR22031315, and SAMN31427486, respectively.
