Figures & data
Figure 1. Plant image of L. carnosum var. drymoglossoides. Sterile leaves oblong or ovate, rounded or obtusely rounded, base cuneate, entire. Fertile leaves ligulate or oblanceolate, narrowly constricted at the base, sometimes the same shape as the sterile leaves, fleshy, leathery when dry. This photograph was taken by Chong-Jian Zhou in Huangshan City, Anhui Province, and was used with the author’s permission.

Figure 2. Chloroplast genome map of L. carnosum var. drymoglossoides. Genes drawn outside the outer circle are transcribed counterclockwise, and genes drawn inside the outer circle are transcribed clockwise. Genes belonging to diferent functional groups are color-coded. The different colored legends in the bottom left corner indicate genes with different functions. The dark grey inner circle indicates the GC content of the chloroplast genome and the presence of nodes in the LSC, SSC, IR regions.
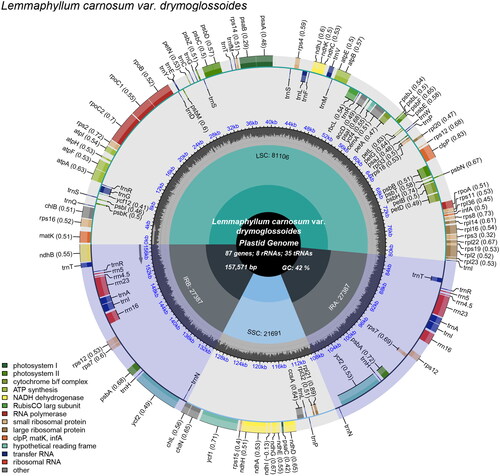
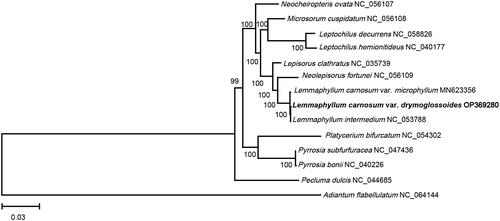
Figure 3. The phylogenetic position for L. carnosum var. drymoglossoides according to the ML phylogenetic tree constructed based on 14 chloroplast genomes. The following sequences were used: Lemmaphyllum carnosum var. drymoglossoides OP369280, Adiantum flabellulatum NC_064144, Lemmaphyllum carnosum var. microphyllum MN623356 (Liu et al. Citation2020), Lepisorus clathratus NC_035739 (Wei et al. Citation2017), Leptochilus decurrens NC_058826 (Su et al. Citation2019), Leptochilus hemionitideus NC_040177 (Min et al. Citation2018), Lemmaphyllum intermedium NC_053788 (Wang et al. Citation2021), Microsorum cuspidatum NC_056108, Neolepisorus fortunei NC_056109, Neocheiropteris ovata NC_056107 (Liu et al. Citation2021), Pecluma dulcis NC_044685 (Samuli and Cárdenas Citation2019), Platycerium bifurcatum NC_054302, Pyrrosia bonii NC_040226 (Cai et al. Citation2018), Pyrrosia subfurfuracea NC_047436 (Min et al. Citation2019). The sequences used for the tree structure are coding sequences. The bootstrap support values are shown on the nodes.
Supplemental Material
Download TIFF Image (273.3 KB)Supplemental Material
Download TIFF Image (150.5 KB)Supplemental Material
Download MS Word (376.3 KB)Data availability statement
The genom esequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OP369280. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA682118, SRR23238769, and SAMN32925244, respectively.
