Figures & data
Figure 1. The morphological characteristics of Eurya rubiginosa var. attenuata. A, B, and C show photos of the whole plant, leaves, and fruits, respectively (photos taken by Yingshuo Li in Qingyuan County, Zhejiang Province, China)

Figure 2. Schematic map of the overall features of the Eurya rubiginosa var. attenuata chloroplast genome. The circular map of the chloroplast genome was generated using CPGview. The map contains six tracks. From the center going outward, the first circle shows the distributed repeats connected with red (the forward direction) and green (the reverse direction) arcs. The second circle shows the long tandem repeats as short blue bars. The third track shows the microsatellite sequences as short bars with different colors. The fourth circle shows the sizes of the chloroplast genome regions, including the small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions. The LSC and SSC regions measure 87255 bp and 18216 bp, respectively, while the inverted repeat regions (IRa and IRb) are both 25872 bp in length. The fifth track shows the GC content along the genome. The sixth circle shows the genes with different colors based on their functional groups. The transcription directions for the inner and outer genes are clockwise and counterclockwise, respectively.
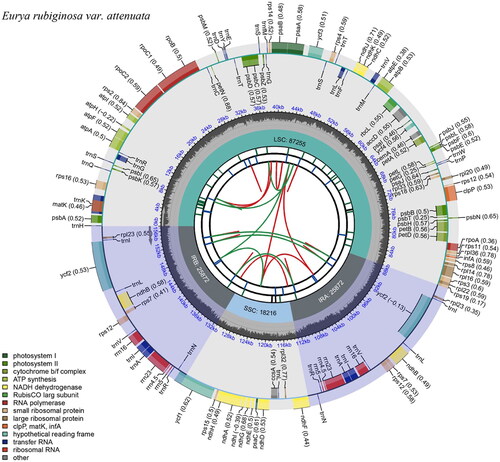
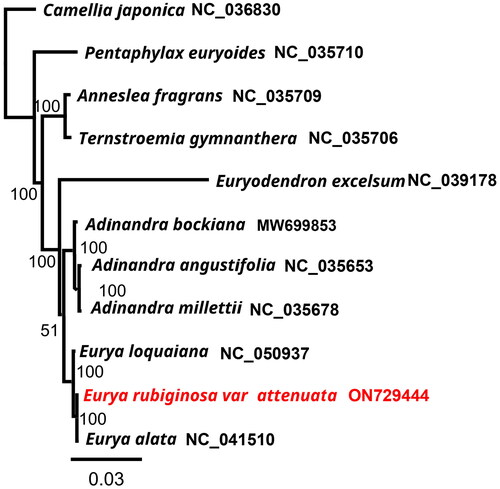
Figure 3. Tree showing the phylogenetic relationships of Eurya rubiginosa var. attenuata. The numbers on the branches represent the bootstrap values based on 1000 replicates. The sequences used for tree construction are as follows: Camellia japonica (NC_036830), Pentaphylax euryoides (NC_035710; Yu et al. Citation2017), Anneslea fragrans (NC_035709; Yu et al. Citation2017), Ternstroemia gymnanthera (NC_035706; Yu et al. Citation2017), Euryodendron excelsum (NC_039178; Shi et al. Citation2019), Adinandra bockiana (MW699853), A. angustifolia (NC_035653; Yu et al. Citation2017), A. millettii (NC_035678; Yu et al. Citation2017), Eurya loquaiana (NC_050937; Wang et al. Citation2020), E. rubiginosa var. attenuata (ON729444), and E. alata (NC_041510; Lin et al. Citation2019).
Supplemental Material
Download MS Word (505.5 KB)Supplemental Material
Download TIFF Image (3.8 MB)Supplemental Material
Download JPEG Image (109 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under accession no. ON729444. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA905216, SRR22406071 and SAMN31867763, respectively.
