Figures & data
Figure 1. Reference image of an adult female Raillietiella orientalis. This image is original work provided by the authors.
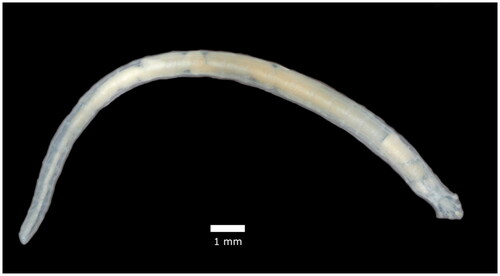
Figure 2. Map of the assembled and annotated Raillietiella orientalis mitochondrial genome (NCBI accession: OP857516). the 13 protein-coding genes, 22 tRNA genes, and two ribosomal RNA genes are shown as colored blocks according to the legend provided. The asterisk for trnC-UAA indicates that this tRNA annotation is incomplete. The grey inner circle represents the local GC content and the blue box highlights a putative at-rich, non-coding control region. The mitogenome map was created using OGDraw v1.3.1 (Greiner et al. Citation2019).
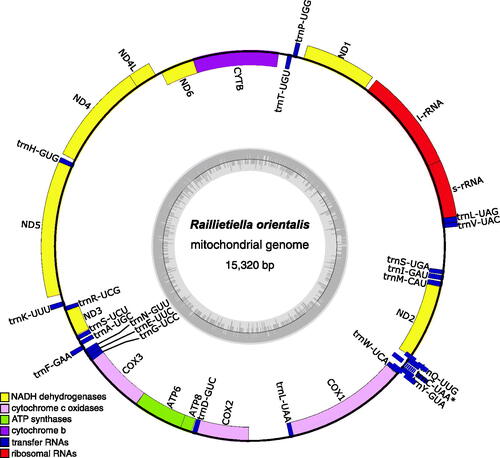
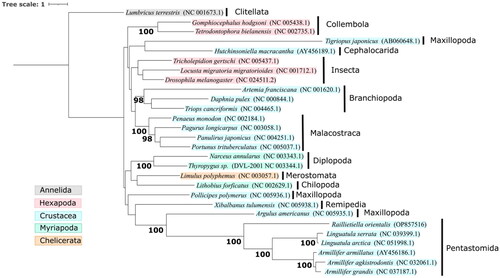
Figure 3. Maximum-likelihood phylogenetic tree based on 13 protein-coding genes from 27 Pancrustacean mitogenomes and 1000 ultrafast bootstrap replicates. Accession numbers are shown within parentheses, and the taxonomic classes are shown by vertical black bars. The Annelid Lumbricus terrestris was used as the outgroup. Nodes with ultrafast bootstrap support ≥95 are labeled.
Data availability statement
The mitogenome sequence assembly is available in NCBI (https://www.ncbi.nlm.nih.gov/) as accession OP857516. The associated BioProject, BioSample, and SRA accessions are PRJNA885072, SAMN31441700, and SRR22081628, respectively.
