Figures & data
Figure 1. Species reference images of Andreaea regularis; whole plant (a), a shoot (b) and a leaf (c). The pictures were taken by authors in a field on Barton Peninsula of King George Island, Antarctica (a) and in a laboratory (b, c). Scale bar = 1 mm (b) and 0.1 mm (c).

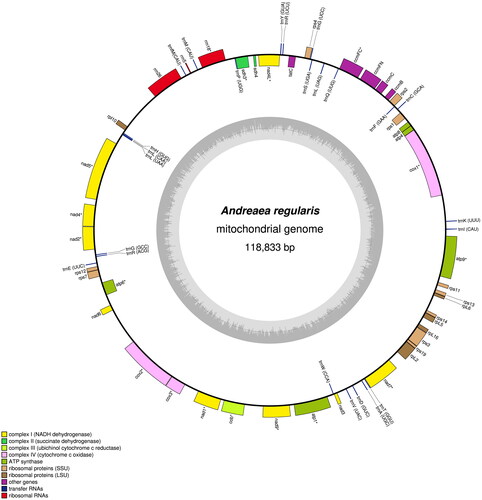
Figure 2. Circular map of the Andreaea regularis mitochondrial genome. Genes outside the outer circle are transcribed clockwise, and those inside the circle are transcribed counterclockwise. Genes belonging to different functional groups are color coded. The innermost darker gray corresponds to GC content, and the lighter gray corresponds to at content.
Supplemental Material
Download Zip (1.1 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. OP067134. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA911323, SRP412618, and SAMN32178634, respectively.