Figures & data
Figure 1. P. amethystina subsp. argutidens species reference image was taken in Shangri-la, Yunan Province, China (27°37′11″N, 99°38′29″E) and provided by corresponding author Huang Yuan. It is a perennial herb with rich violet-blue flowers and deeply serrated leaves.

Figure 2. Genomic map of overall features of Primula amethystina subsp. argutidens chloroplast genome, generated by CPGview (http://www.1kmpg.cn/cpgview/). The species’ name is shown in the top left corner. The map contains six tracks by default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
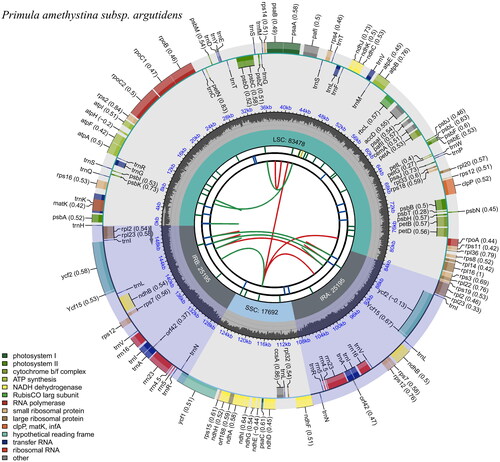
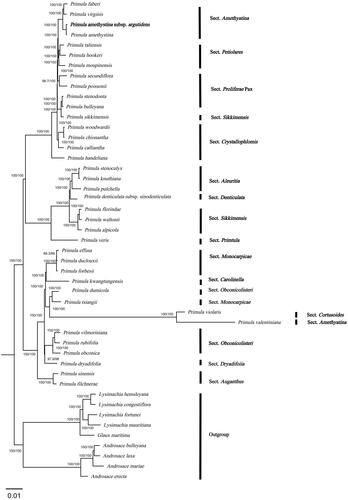
Figure 3. In the ML phylogenetic tree of P. amethystina subsp. argutidens and 46 Primulaceae species based on chloroplast genomes. The best-fit model according to the Bayesian information criterion (BIC) was TVM + F + R3. Branch supports were tested using ultrafast bootstrap (UFBoot) and SH-like approximate likelihood ratio test (SH-aLRT) with 10,000 replicates. P. amethystina subsp. argutidens is marked in bold. The following sequences were used: Primula faberi NC_053576 (Wang et al. Citation2021), Primula virginis NC_053581 (Wang et al. Citation2021), Primula amethystina NC_053577 (Wang et al. Citation2021), Primula taliensis NC_053601 (Wang et al. Citation2021), Primula hookeri NC_053593 (Wang et al. Citation2021), Primula moupinensis NC_050244, Primula secundiflora NC 053585 (Wang et al. Citation2021), Primula poissonii NC_024543 (Yang et al. Citation2014), Primula stenodonta NC 034677 (Zhang et al. Citation2017), Primula bulleyana NC_046947 (Chen, Zhang et al. Citation2019), Primula sikkimensis NC_050243, Primula woodwardii NC 039349 (Ren et al. Citation2018), Primula chionantha NC_053583 (Wang et al. Citation2021), Primula calliantha MZ054238 (Yang et al. Citation2021), Primula handeliana NC_039348 (Ren et al. Citation2018), Primula stenocalyx NC_058249 (Guo, Ma, Li et al. Citation2021), Primula knuthiana NC_039350 (Ren et al. Citation2018), Primula pulchella NC_050246 (Zhang et al. Citation2017), Primula denticulata subsp. sinodenticulata NC_050247, Primula florindae NC_053579 (Wang et al. Citation2021), Primula waltonii NC_058808, Primula alpicola NC_053588 (Wang et al. Citation2021), Primula veris NC_031428 (Zhou et al. Citation2016), Primula effusa NC_058259 (Zhong et al. Citation2019), Primula duclouxii NC_058263 (Zhong et al. Citation2019), Primula forbesii NC_061696, Primula kwangtungensis NC_034371 (Zhang et al. Citation2016), Primula dumicola NC_058257 (Zhong et al. Citation2019), Primula tsiangii NC_046755 (Chen, Yan et al. Citation2019), Primula violaris NC_050848 (Chai et al. Citation2021), Primula valentiniana NC_061669, Primula vilmoriniana NC_058258 (Zhong et al. Citation2019), Primula rubifolia NC_058261 (Zhong et al. Citation2019), Primula obconica NC_046415 (Zhang et al. Citation2019), Primula dryadifolia NC_053596 (Wang et al. Citation2021), Primula sinensis NC_030609 (Liu et al. Citation2016), Primula filchnerae NC_051972 (Lu et al. Citation2020), Lysimachia hemsleyana NC_052863 (Ying et al. Citation2019), Lysimachia congestiflora NC_045275 (Li et al. Citation2019), Lysimachia fortunei NC_052863, Lysimachia mauritiana NC_060700 (Lee et al. Citation2022), Glaux maritima NC_059901 (Liu et al. Citation2021), Androsace bulleyana NC_034641, Androsace laxa (Ren et al. Citation2018), Androsace mariae NC_051991 (Guo, Ma, Xie et al. Citation2021), and Androsace erecta NC 057637.
Supplemental Material
Download MS Word (454.9 KB)Data availability statement
The genome sequence data supporting the study’s findings are openly available in NCBI GenBank at https://www.ncbi.nlm.nih.gov/ under accession no. ON416902. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA834988, SRR19175835, and SAMN28088488, respectively.
