Figures & data
Figure 1. The azolla-algae community and the algal cells. (A) Azolla-algae community; (B) wild algal cells from the community; algal cells grown in TAP medium and stained with Nile red, and observed under bright (C) and fluorescent light (D). Red arrow indicates the algal film in the gap between azolla plants; blue arrows indicate the oil droplets in the algal cells. (A, B) Taken by Jiaming Zhang; (C, D) taken by Yaojia Mu.
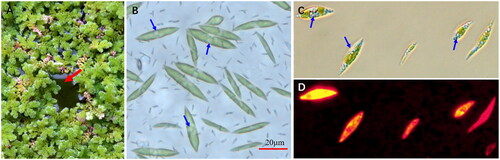
Figure 2. Organization of the chloroplast genome of C. braunii ITBB-AG6. (A) Coverage depth plot of the genome, the depth was normalized using the logarithm based on 2. (B) Circular map of the genome with genes’ location, size, and orientation indicated by blue (CDS), green (rRNA), and red (tRNA) arrows.
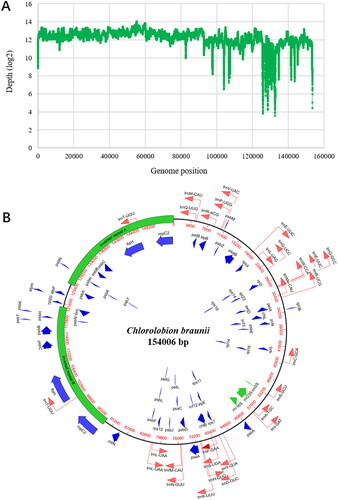
Table 1. Gene list of the chloroplast genome of C. braunii ITBB-AG6.
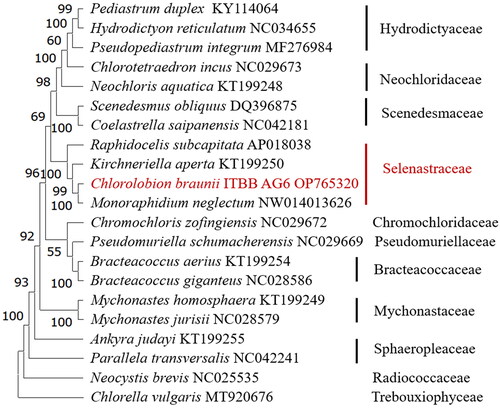
Figure 3. Consensus maximum-likelihood tree of Sphaeropleales (chlorophyta). The tree is rooted with a Chlorella vulgaris genome (Han et al. Citation2021). Bootstrap supports of 1000 replicates are shown above the branches. The strain in this study is shown in red.
Data availability statement
The complete chloroplast genome sequence is openly available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. OP765320. The Illumina reads generated by the authors to assemble this genome are available under GenBank BioProject no. PRJNA931121, BioSample no. SAMN33050851, and SRA no. SRR23329251.
