Figures & data
Figure 1. Plant characteristics image of Cuminum cyminum. (A) the C. cyminum cultivated in Tulufan city, Xinjiang province, China. It is between 50 and 120 cm high and racemes termina; stem is erect, upright part is quadrangular, the edge is densely covered with white hairs, the lower and Middle branches are often purple; leaves are rounded kidney-shaped, leaves green glabrous or pubescent, grey-green below pubescent. (B) the C. cyminum matures, dries and turns yellow in the plantation at june. (C) the C. cyminum seeds are ellipsoidal or quasi-circular in shape, 5–8 mm in length, grayish white to dark grey in surface, with irregular longitudinal projections and striations, shiny. (D) the germination of seed tissue culture to form aseptic seedling was used to extract DNA in this study. The photograph A, and B were taken by the Feiya Suo in the Tulufan, Xinjiang province, China (89°1857′N, 42°6793′W). the photograph C, and D was taken by the Luodong Huang on 10 october 2022 at the biological resources and environmental microbiology laboratory of the College of Life Science and Technology,Guangxi University.

Figure 2. Mitogenome circular map of Cuminum cyminum. The genes name on the outside line side indicated that these genes were located on the H-strand, whereas the others were located on the L-strand. Genes are shown in different color blocks on the map.
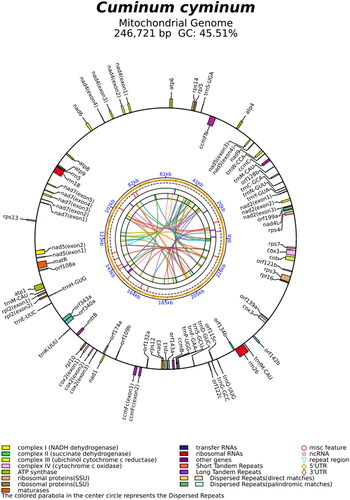
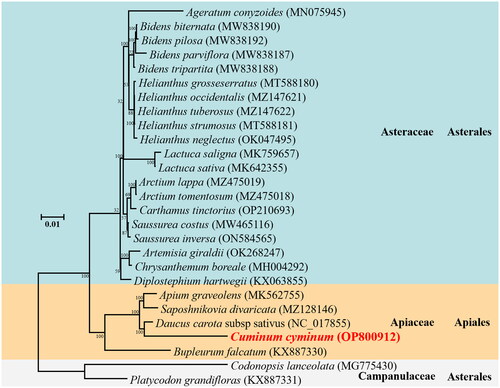
Figure 3. Phylogenetic relationships for Cuminum cyminum according to the maximum-likelihood phylogenetic tree constructed based on 27 mitogenomes of campanulids species. Node labels indicate bootstrap values. GenBank accession numbers of the following sequences were used: Ageratum conyzoides (MN075945) (Luo and Pan Citation2019), Bidens biternata (MW838190) (Yao et al. 2022#), Bidens pilosa (MW838192) (Yao et al. 2022#), Bidens parviflora (MW838187) (Yao et al. 2022#), Bidens tripartita (MW838188) (Yao et al. 2022#), Helianthus grosseserratus (MT588180) (Makarenko et al. Citation2020), Helianthus occidentalis (MZ147621) (Makarenko et al. Citation2021), Helianthus tuberosus (MZ147622) (Makarenko et al. Citation2021), Helianthus strumosus (MT588181) (Makarenko et al. Citation2020), Helianthus neglectus (OK047495) (Khachumov et al. 2021#), Lactuca saligna (MK759657) (Kozik et al. Citation2019), Lactuca sativa (MK642355) (Kozik et al. Citation2019), Arctium lappa (MZ475019) (Xu and Kang 2021#), Arctium tomentosum (MZ475018) (Xu and Kang 2021#), Carthamus tinctorius (OP210693) (Yang 2019#), Saussurea costus (MW465116) (Song et al. Citation2021), Saussurea inversa (ON584565) (Dai et al. 2022#), Artemisia giraldii (OK268247) (Yue 2022#), Chrysanthemum boreale (MH004292) (Won et al. Citation2018), Diplostephium hartwegii (KX063855) (Vargas et al. Citation2017), Apium graveolens (MK562755) (Cheng et al. Citation2021), Saposhnikovia divaricata (MZ128146) (Ni 2021#), Daucus carota subsp sativus (NC_017855) (Iorizzo et al. Citation2012), Bupleurum falcatum (KX887330) (Kim et al. Citation2020), Codonopsis lanceolata (MG775430) (Lee et al. Citation2018), Platycodon grandifloras (KX887331) (Lee et al. Citation2018). #direct submission to NCBI, unpublished.
Supplemental Material
Download PDF (1.4 MB)Supplemental Material
Download JPEG Image (1 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI under accession no. OP800912. The associated BioProject, BioSample, and SRA numbers are PRJNA899008, SAMN31636213, and SRR22214714, respectively.
