Figures & data
Figure 1. Images of Aegle marmelos (L.) Correa 1800: (a) Leaves; (b) fruit; (c) whole plant. A. marmelos is a deciduous small tree, approximately 10 m tall, with leaflets featuring numerous oil glandular dots. The leaflets are broadly ovate or long elliptic, measuring 4–12 cm in length. terminal leaflets are larger and long-stalked, while lateral leaflets are subsessile with shallowly obtuse serrations. The fruit measures 10–12 cm in height and 6–8 cm in diameter, displaying a light greenish-yellow color, smooth texture, and a hard woody pericarp, approximately 3–4 mm thick. The fruit stalk is 4–6 cm long, and the seeds, numerous in quantity, are compressed ovoid with transparent gum. The testa is wooly, and cotyledons emerge during germination. These photos were captured by a local professional team in Xishuangbanna County, Yunnan Province, China.

Figure 2. Chloroplast reference genome of Aegle marmelos. The diagram illustrates the organization of the chloroplast genome. Starting from the center, the first circle represents the forward and reverse repeat sequences, indicated by red and green arcs, respectively. The second circle depicts a tandem repeat sequence, while the third circle highlights microsatellite sequences with short strips. The fourth circle displays the positions of the large single-copy gene region (LSC), small single-copy gene region (SSC), and inverted repeat regions (IRA and IRB). the fifth circle represents the GC content. The genes located outside the sixth circle are transcribed counterclockwise, while those inside are transcribed clockwise. Different functional groups of genes are color-coded, as indicated in the legend.
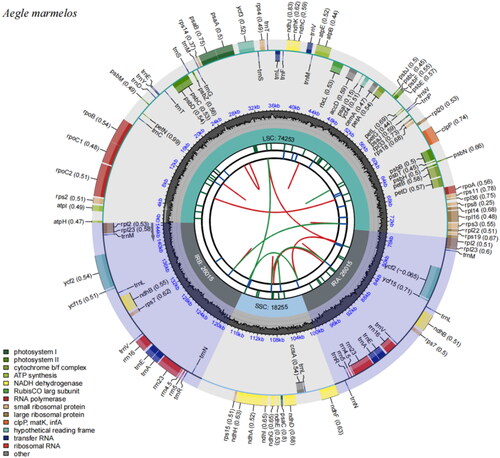
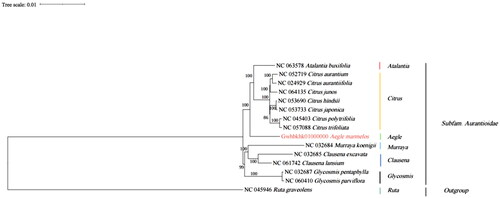
Figure 3. Showcases the phylogenetic analysis of 14 species and 1 taxonomic unit outgroup, utilizing the complete chloroplast genome sequence with the RAxML method. The following sequences were used for analysis: NC_063578 Atalantia buxifolia (Wang et al. Citation2017), NC_052719 Citrus aurantium (Lin et al. Citation2020), NC_024929 Citrus aurantiifolia (Su et al. Citation2014), NC_064135 Citrus juno (Zhong-Wang Citation2008), NC_053690 Citrus hindsii (Xu et al. Citation2019), NC_053733 Citrus japonica (Wu et al. Citation2019), NC_045403 Citrus polytrifolia (Li et al. Citation2019), NC_057088 Citrus trifoliata, Gwhbkhk01000000 Aegle marmelos, NC_032684 Murraya koenigii (Shivakumar et al. Citation2017), NC_032685 Clausena excavata (Natthawadi et al. Citation2018), NC_061742 Clausena lansium (Zhao et al. Citation2019), NC_032687 Glycosmis pentaphylla (Shivakumar et al. Citation2017), NC_060410 Glycosmis parviflora (Wen-Fang et al. Citation2013), and NC_045946 Ruta graveolens (Ling and Zhang Citation2019).
Supplemental Material
Download MS Word (197 KB)Data availability statement
The genome sequence data obtained in this study are openly available in GenBank of NGDC (https://ngdc.cncb.ac.cn/) under the accession number Gwhbkhk01000000. The associated BioProject, Bio-Sample and GSA numbers are PRJCA011521, SAMC866117 and CRA007998, respectively.
