Figures & data
Figure 1. Photographs of Crepidomanes latealatum taken by S. H. Park at the sample collection site: (A) C. latealatum growing on a shady rock near a humid valley. (B) Winged rachis and tubular involucres with dilated lips of C. latealatum.

Figure 2. Complete chloroplast genome map of Crepidomanes latealatum, containing six tracks. From the center, the first track shows the dispersed repeats, which consist of direct (red) and palindromic (green) repeats. The second and third tracks show the long and short tandem repeats, respectively. The regional composition of the genome, LSC, SSC, and IRs, are identified on the fourth track. The GC content along the genome is plotted in the fifth track. The genes are shown on the outer sixth track.
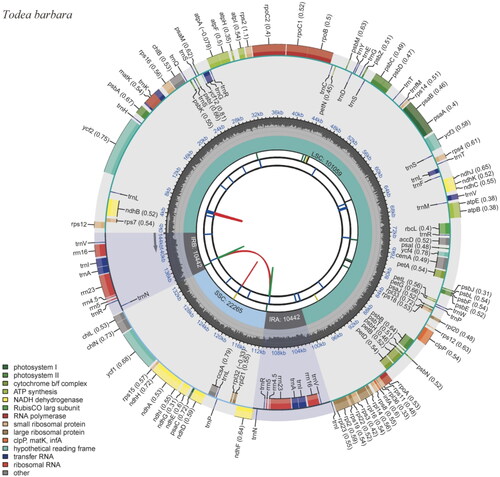
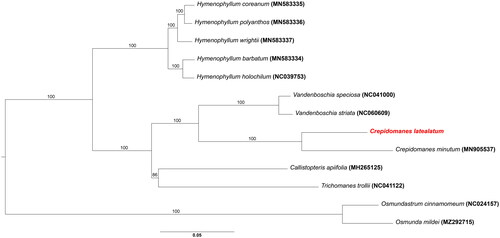
Figure 3. Phylogenetic tree of Crepidomanes latealatum and related taxa inferred from maximum likelihood analysis based on the 88 coding genes sequences. The outgroup was Osmundastrum chinnamomeum (NC 024157; Kim et al. Citation2014) and 5 genera of the family Hymenophyllaceae were included together with osmunda mildei (MZ292715). the bootstrap supporting values are described on the branches. The position of C. latealatum is indicated in red. The following sequences were used: MN583336 for hymenophyllum polyanthos (Kim and Kim Citation2020), MN583335 for H. coreanum (Kim and Kim Citation2020), MN583337 for H. wrightii (Kim and Kim Citation2020), MN039753 for H. holochilum (Kuo et al. Citation2018), NC063573 for Crepidomanes minutum, NC041000 for Vandenboschia speciosa (Ruiz-Ruano et al. Citation2019), NC060609 for V. striata (Wang et al. Citation2022), MH265125 for callistopteris apiifolia, and NC041122 for Trichomanes trollii (Lehtonen Citation2018).
Supplemental Material
Download JPEG Image (2.3 MB)Supplemental Material
Download JPEG Image (7 MB)Data availability statement
The genome sequence data supporting the findings of this study are available in the NCBI GenBank under accession no. NC072291 (https://www.ncbi.nlm.nih.gov/nuccore/ NC_072291.1). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA951456, SRR24044269, and SAMN34043022, respectively.
