Figures & data
Figure 1. Photographs of Todea barbara captured by Ki-Joong Kim at the collection site. The Austral king fern has a barrel-shaped trunk covered in dark brown to black fibrous aerial roots. The stipes are smooth and yellow-brown, with ear-like lobes at the base. The fronds are large and coriaceous.

Figure 2. Complete chloroplast (cp) genome map of Todea barbara, containing six tracks. From the center, the first track shows the dispersed repeats, which consist of direct (red) and palindromic (green) repeats. The second and third tracks show the long and short tandem repeats, respectively. The regional composition of the genome, LSC, SSC, and IRs, are identified on the fourth track. The GC content along the genome is plotted in the fifth track. The genes are shown on the outer sixth track.
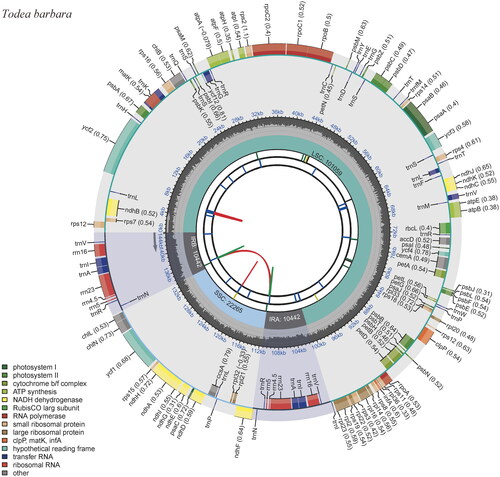
Figure 3. The internal stop codons in eight genes of Todea barbara. The numbers above the bases indicate the positions of the gene.

Table 1. Comparison of the complete chloroplast genomes of Todea and its related species used in the present study.
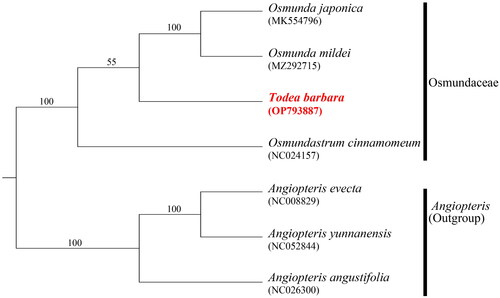
Figure 4. Maximum likelihood tree based on the 85 cp coding genes. Numbers on the nodes represent the bootstrap support values. The outgroups were three Angiopteris of A. evecta (NC024157; Roper et al. Citation2007), A. yunnanensis (NC052844; Jiang et al. Citation2019), and A. angustifolia (NC026300). Two Osumnda species (O. japonica; MK554796, Xu et al. Citation2019 and O. mildei; MZ292715) and Osmundatrum cinnamomeum (NC024157; Kim et al. Citation2014) were also included for the molecular phylogenetic analysis of the family Osmundaceae.
Data availability statement
The genome sequence data supporting the findings of this study are available from NCBI GenBank under accession no. OP793887 (https://www.ncbi.nlm.nih.gov/nuccore/OP793887.1/). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA954512, SRR24137031, and SAMN34146952, respectively.
