Figures & data
Figure 1. Photos of Ceriodaphnia dubia. This species can be distinguished from other related species based on dorsolateral depression between head and rest of body, (A) postadominal claw (arrow), and (B) lack of rostrum and tail spine. The photos were taken by the authors.
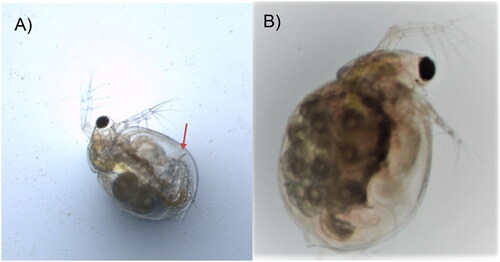
Figure 2. Circular map of the mitochondrial genome of C. dubia. Genes outside the circle are encoded on the heavy strand and genes inside the circle are encoded on the light stand. The inner black bars indicate the GC content, and the Middle line represents 50%. Visualization was performed using Proksee (Grant et al. Citation2023).
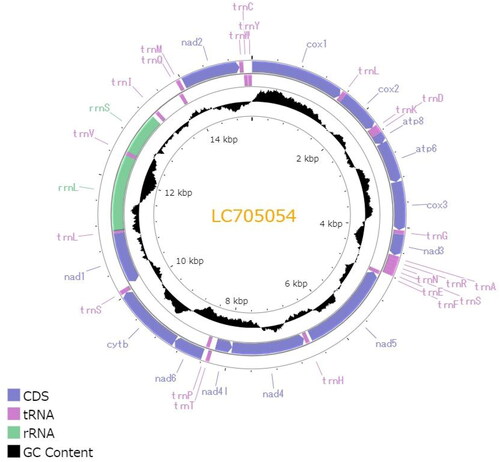
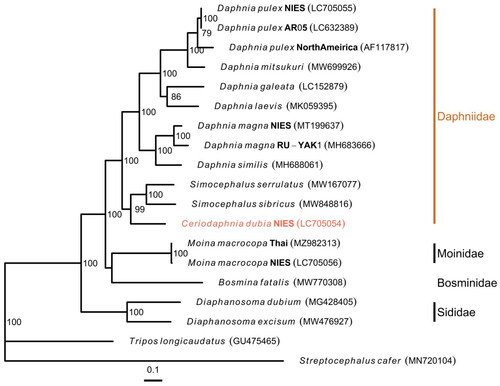
Figure 3. Maximum likelihood Tree based on 13 PCGs in mitochondrial genomes of cladocerans. Red species represent the mitogenomes obtained in this study. S. cafer and T. longicaudatus were used as the outgroup. Non-parametric bootstrap values (based on 3000 times resampling) are shown at nodes. The scale bar indicates the number of amino acid substitutions per site. The following sequences were used to infer the tree: Daphnia pulex LC705055.1, D. pulex LC632389.1 (Ohtsuki et al. Citation2022), D. pulex AF117817.1 (Crease Citation1999), D. mitsukuri MW699926.1, D. galeata LC152879.1 (Tokishita et al. Citation2017), D. laevis MK059395.1 (Martins Ribeiro et al. Citation2019), D. magna MT199637.1 (Lee et al. Citation2020), D. magna MH683666.1 (Fields et al. Citation2018), D. similus MH688061.1 (Fields et al. Citation2018), Simocephalus serrulatus MW167077.1, S. sibricus MW848816.1 (Gu et al. Citation2021), C. dubia LC705054.2 (this study), Moina macrocopa MZ982313.1 (Nam et al. Citation2022), M. macrocopa LC705056.1, bosmina fatalis MW770308.1 (Wei et al. Citation2021), Diaphanosoma dubium MG428405.1 (Liu et al. Citation2017), Diaphanosoma excisum MW476927.1 (Pan et al. Citation2021), tripos longicaudatus GU475465.1 (Ryu and Hwang Citation2010), and streptocephalus cafer MN720104.1 (Tladi et al. Citation2020).
Supplemental Material
Download MS Word (110.4 KB)Data availability statement
Data that support the findings of this study are openly available at INSDC with the accession number LC705054.2, and at Sequence Read Archives (SRA) with the accession number PRJDB13090 (BioProject), SAMD00444315 (C. dubia) (BioSample), DRX338201 and DRX338203 (Experiment), and DRR352290 and DRR352292 (Run). The sequences at two different SRA accession numbers were derived from the same DNA sample but from different sequencing runs.
