Figures & data
Figure 1. The individual of S. braziliana used in this study. Its morphotype is similar to the one shown in Figure 4(E) in the previous taxonomic study (Carmona et al. Citation2014). The picture was taken by Hideaki Mizobata.

Table 1. The list of species, GenBank accession ID, and references used for alignments (Supplementary Data 1).
Figure 2. Circular map of the complete mitochondrial genome annotated to show the locations and orientation of its 13 protein-coding genes, 2 rRNA genes, and 23 tRNA genes. The upper arrows indicate the direction of transcription, while the inner gray circle represents the GC content.
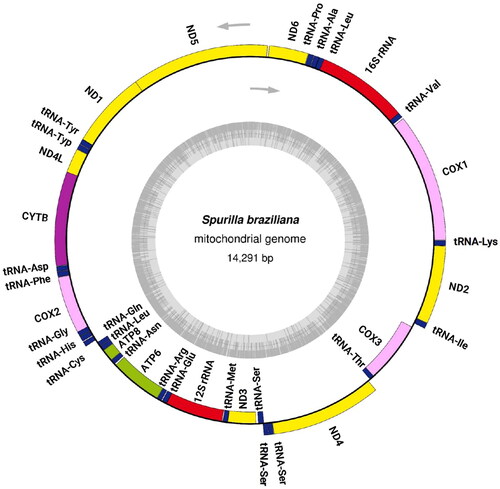
Figure 3. Maximum likelihood phylogenetic tree of S. braziliana and six other Cladobranchia species based on the concatenated amino acid alignments of the 13 protein-coding genes in the mitochondrial genome. Bootstrap support values are shown for each node, based on 1000 replicates. The list of species and their GenBank accession IDs are shown in .

Supplemental Material
Download PNG Image (133.4 KB)Supplemental Material
Download MS Power Point (117.6 KB)Supplemental Material
Download MS Power Point (1.2 MB)Supplemental Material
Download MS Power Point (10.5 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession ID: LC759638. The associated BioProject, SRA, and Bio-sample numbers are PRJDB15401, DRR450785, and SAMD00585630, respectively.
