Figures & data
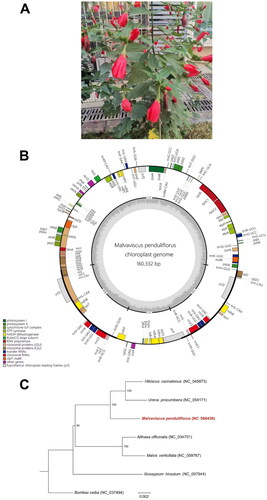
Figure 1. (A) an Individual plant of M. penduliflorus taken from Chenghua District, Chengdu, China (photoed by Dr. Rong Liu). (B) The detailed genome map of M. penduliflorus cp genome. The large single copy (LSC), small single copy (SSC) region and two inverted repeat regions (IRA and IRB), and GC content (light gray) are shown in the inside track. Gene models including protein-coding genes, tRNA genes, and rRNA genes are shown with various colored boxes in the outer track. (C) Maximum likelihood species tree based on the coding sequences of 75 genes that are presented in all the six cp genomes of Malvaceae species, including M. penduliflorus, U. procumbens (Wang et al. Citation2021), H. cannabinus (Cheng et al. Citation2020), A. officinalis (Zhang et al. Citation2017), M. verticillata (Li et al. Citation2020), and G. hirsutum (Lee et al. Citation2006), using B. ceiba (Gao et al. Citation2018) as outgroup. Bootstrap support values were denoted on each internal node, indicating that M. penduliflorus clustered with U. procumbens and H. cannabinus with 100% bootstrap support.
Supplemental Material
Download MS Word (109 KB)Data availability statement
The genome sequence data supporting the findings of this study are openly available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession no. NC_066439. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA885282, SRR21748890, and SAMN31083560, respectively.
