Figures & data
Figure 1. An individual of Salix lindleyana (species reference map was taken by Xin Du and Xiu-Xing zhangin Kangding City, Ganzi Tibetan Autonomous Prefecture, Sichuan Province, China (29°54′18″N, 102°0′7″E). Salix lindleyana is a kind of cushion shrub, which stem prostrate and rooting. Leaves obovate-oblong or obovate-lanceolate, bright green above, glabrous, midrib distinctly concave. Inflorescences ovate, with only a few flowers per inflorescence, born at the tip of the current branch. Bracts broadly ovate, apex rounded, pale purplish-red.

Figure 2. Schematic representation of the plastome features of Salix lindleyana. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. Black: c (complex repeat); green: p1 (repeat unit size = 1); yellow: p2 (repeat unit size = 2); purple: p3 (repeat unit size = 3); blue: p4 (repeat unit size = 4); orange: p5 (repeat unit size = 5); red: p6 (repeat unit size = 6). the small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The base frequency at each site along the genome will be shown between the fourth and fifth tracks. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
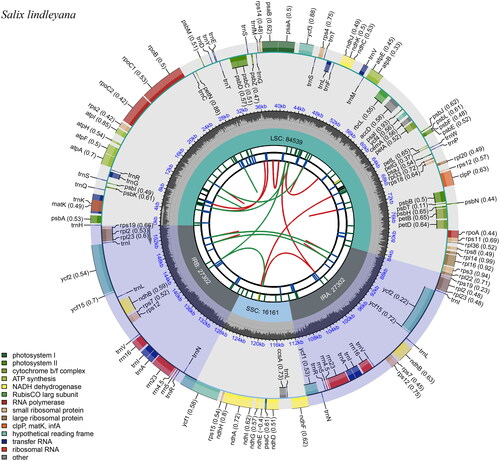
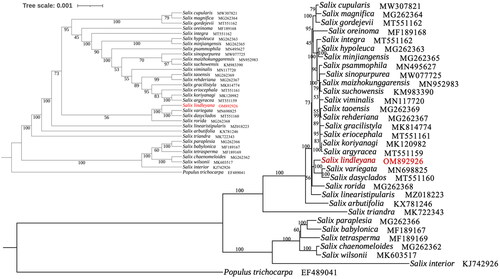
Figure 3. Phylogenetic tree of 32Salix species was established with maximum likelihood using the complete chloroplast genome. The cladogram tree was placed in the upper left corner. Number on each node indicated bootstrap support value. The following sequences were used: Salix cupularis (MW307821; Li et al. Citation2021), Salix magnifica (MG262364; Liu et al. Citation2020), Salix gordejevii (MW562004; Wei and Li Citation2021), Salix oreinoma (MF189168; Huang et al. Citation2017), Salix integra (MT551162; Zhou et al. Citation2021), Salix hypoleuca (MG262363; unpublished), Salix minjiangensis (MG262365; unpublished), Salix psammophila (MN495627; Lu et al. Citation2019), Salix sinopurpurea (MW077725; Guo et al. Citation2021), Salix maizhokunggarensis (MN952983; Ma et al. Citation2020), Salix suchowensis (MG262365; Wu Citation2016), Salix viminalis (MN117720; Hu et al. Citation2019), Salix taoensis (MG262369; unpublished), Salix rehderiana (NC_037427; unpublished), Salix gracilistyla (MK814774; Xi et al. Citation2019), Salix eriocephala (MT551161; unpublished), Salix koriyanagi (MK120982; Kim et al. Citation2019), Salix argyracea (MT551159; unpublished), Salix variegate (MN698825; Chen Citation2020), Salix dasyclados (MT551160; unpublished), Salix rorida (MG262368; unpublished), Salix linearistipularis (MZ018223; ren et al. 2021), Salix arbutifolia (KX781246; unpublished), Salix triandra (MK722343; Wu et al. Citation2019(a)), Salix paraplesia (MG262366; unpublished), Salix babylonica (MF189167; Huang et al. Citation2017), Salix tetrasperma (MF189169; Huang et al. Citation2017), Salix chaenomeloides (MG262362; unpublished), Salix wilsonii (MK603517; Wu et al. Citation2019(b)), Salix interior (KJ742926; Huang et al. Citation2014).
Supplemental Material
Download JPEG Image (133 KB)Supplemental Material
Download JPEG Image (276.3 KB)Supplemental Material
Download JPEG Image (1.8 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number OM892926. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA835694, SRR19116050, and SAMN28108208 respectively.
