Figures & data
Figure 1. The species image of adult E. godlewskii. This bird is characterized by a grey head with chestnut lateral stripe, which is widely distributed in East Asia and tends to select bushy and rocky hill slopes that are often near forests, thickets, ravines, and farm fields. The photograph was taken in Jinzhong mountain nature reserve in Guangxi province of China by Dr X.Bao.

Figure 2. The circular complete mitochondrial genome map of E. godlewskii. The complete mtDNA contains 13 PCGs, 23 tRNA genes, 2 rRNA genes (12S rRNA and 16S rRNA), and a control region. The 13 PCGs (ND1, ND2, COX1, COX2, ATP8, ATP6, COX3, ND3, ND4L, ND4, ND5, CYTB and ND6), 23 tRNA genes, and 2 rRNA genes are encoded in the plus and negative strands of the mtDNA. The control region of the mtDNA distributes between genes of tRNA-Glu and tRNA-Phe. The outside of the ring represents the plus strand, while the inside represents the negative strand.
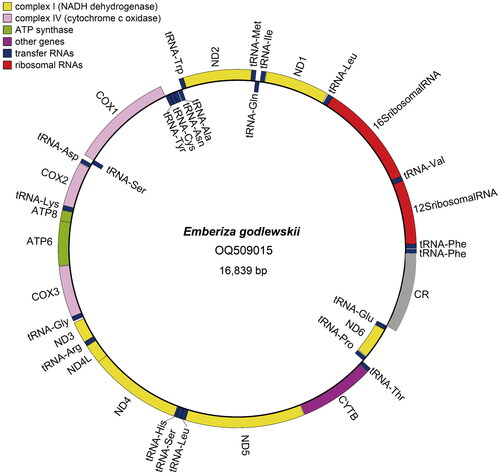
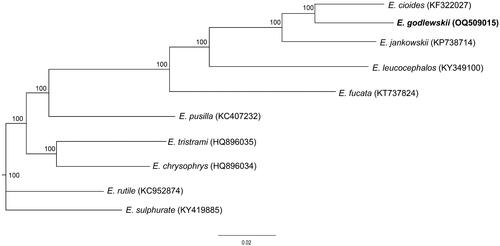
Figure 3. ML tree showing phylogenetic relationship for ten species of genus Emberiza based on the concatenated nucleotide sequences of 13 PCGs with HKY + I + G model. Numbers above or below nodes indicated the bootstrap support values estimated with 1000 replicates. The following sequences were used: Emberiza cioides, NC_024524, KF322027; E. jankowskii, NC_027251, KP738714; E. leucocephalos, NC_037692, KY349100; E. fucata, NC_033338, KT737824; E. pusilla, NC_021408, KC407232; E. tristrami, NC_015234, HQ896035; E. chrysophrys, NC_015233, HQ896034; E. rutile, NC_024925, KC952874; E. sulfurate, KY419885. The support values are shown next to the nodes. The species under study is bolded.
Supplemental Material
Download JPEG Image (502.3 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov under the accession number OQ509015. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA940058, SRR23680762, and SAMN33550910, respectively.
