Figures & data
Figure 1. Light Micrograph of Coelastrum microporum, taken by the authors, from the strain deposited as UTEX 3178.
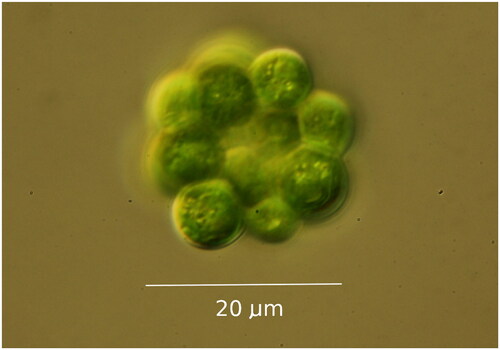
Figure 2. Gene map of Coelastrum microporum plastome. Inverted repeats (IRA and IRB) and two single-copy regions are indicated on the inner circle with G/C content (dark grey) and A/T content (light grey). colored gene boxes are indicated by functional group as shown in the key.
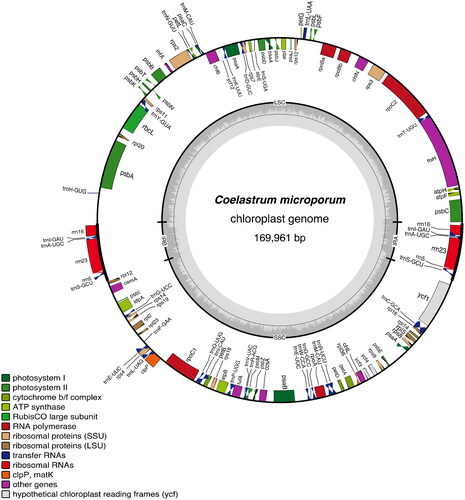
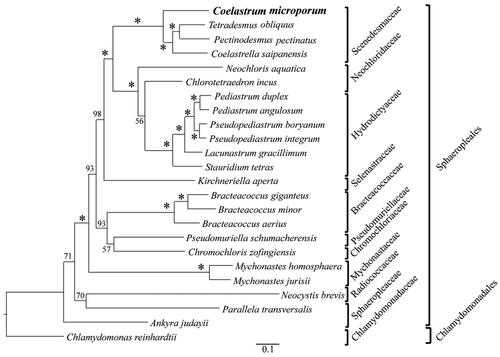
Figure 3. Maximum likelihood phylogeny of Coelastrum microporum, 22 species in Sphaeropleales and Chlamydomonas reinhardtii (outgroup) based on 58 CDSs shared by C. microporum and 23 publicly accessible plastomes in Sphaeropleales and chlamydomonadales. Numbers above branches are bootstrap values from 1000 bootstrap replicates in which asterisks represent bootstrap values of 100. The following sequences were used: Tetradesmus obliquus NC_008101 (de Cambiaire et al. Citation2006), Pectinodesmus pectinatus NC_036668 (unpublished), Coelastrella saipanensis NC_042181 (unpublished), neochloris aquatica NC_029670 (Fučíková et al. Citation2016), chlorotetraedron incus NC_029673 (Fučíková et al. Citation2016), Pediastrum duplex NC_034654 (McManus et al. Citation2017), Pediastrum angulosum NC_037919 (McManus et al. Citation2018), Pseudopediastrum boryanum NC_037920 (McManus et al. Citation2018), Pseudopediastrum integrum NC_037921 (McManus et al. Citation2018), lacunastrum gracillimum NC_037918 (McManus et al. Citation2018), stauridium tetras NC_037923 (McManus et al. Citation2018), kirchneriella aperta NC_029676 (Fučíková et al. Citation2016), Bracteacoccus giganteus NC_028586 (Lemieux et al. Citation2015), Bracteacoccus minor NC_029674 (Fučíková et al. Citation2016), Bracteacoccus aerius NC_029675 (Fučíková et al. Citation2016), pseudomuriella schumacherensis NC_029669 (Fučíková et al. Citation2016), chromochloris zofingiensis NC_029672 (Fučíková et al. Citation2016), Mychonastes homosphaera NC_029671 (Fučíková et al. Citation2016), Mychonastes jurisii NC_028579 (Lemieux et al. Citation2015), neocystis brevis NC_025535 (Lemieux et al. Citation2014), parallela transversalis NC_042241 (unpublished), ankyra judayi NC_029735 (Fučíková et al. Citation2016), Chlamydomonas reinhardtii NC_005353 (Maul et al. Citation2002).
Supplemental Material
Download MS Word (87.4 KB)Data availability statement
The genome sequence data that supported the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. NC_068582. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA909268, PRJNA909268, and SAMN32062636, respectively. The strain used is available from the UTEX Culture Collection of Algae (https://utex.org, Dr. David Nobles, Curator and Director: [email protected]) as Coelastrum microporum UTEX LB 3178.
