Figures & data
Figure 1. The photograph of Castanopsis hystrix. This image was taken by Hui Zhu at Guangxi Forestry Research Institute, Nanning Guangxi, China. C. hystrix is an evergreen broad-leaved tree with leaf blade margin shallowly, serrate from middle to apex, secondary veins usually not reaching margin and petiole rarely longer than 1 cm.

Figure 2. The complete chloroplast genome map of Castanopsis hystrix, which was generated by CPGview. LSC, SSC, and IR (IRa and IRb) with their length are represented on the first circle. The second circle shows the GC ratio in dark gray. The third circle displays the genes with the colors based on their functional classification presented at the left of the circular map. Genes located on the inner are transcribed in a clockwise, and those outer of circle are transcribed in an anticlockwise.
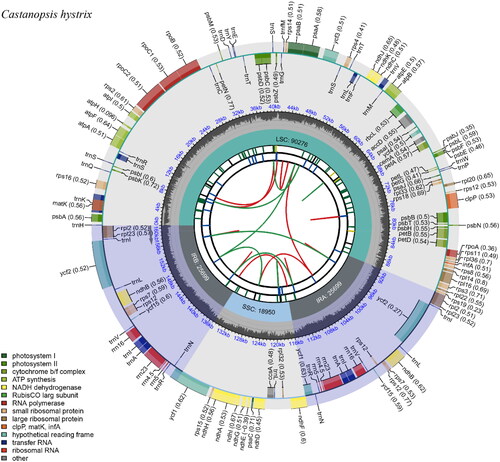
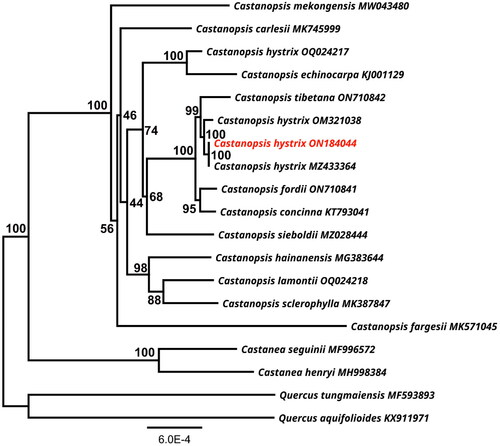
Figure 3. A maximum-likelihood (ML) based phylogenetic tree of Castanopsis hystrix using Quercus aquifolioides and Q. tungmaiensis as outgroups. The scale bar represents the number of nucleotide substitutions per site. The numbers on each node indicated the ML bootstrap support with 1000 replicates. The following sequences were used: Castanea seguinii MF996572, Castanea henryi MH998384 (Gao et al. Citation2019), Castanopsis carlesii MK745999 (Sun et al. Citation2019), Castanopsis concinna KT793041, Castanopsis echinocarpa KJ001129, Castanopsis fargesii MK571045 (Ye et al. Citation2019), Castanopsis fordii ON710841 (Wang et al. Citation2023), Castanopsis hystrix OQ024217, Castanopsis hystrix ON184044 (this study), Castanopsis hystrix OM321038, Castanopsis hystrix MZ433364, Castanopsis sieboldii MZ028444 (Park et al. Citation2021), Castanopsis hainanensis MG383644 (Chen et al. Citation2018), Castanopsis mekongensis MW043480 (Peng et al. Citation2021), Castanopsis sclerophylla MK387847 (Ye et al. Citation2019), Castanopsis tibetana ON710842, Quercus aquifolioides KX911971 (Yang et al. Citation2018), Quercus tungmaiensis MF593893 (Yang et al. Citation2018).
Supplemental Material
Download MS Word (503 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession number ON184044. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA828973, SRR18843872 and SAMN27681347, respectively.
