Figures & data
Figure 1. External morphology of Paracondylactis sinensis, showing its smooth column and oral disk with about 96 tentacles (specimen no. ss-Taizhou-1). The specimen was photographed by Yang Li.

Figure 2. A circular genomic map of the mitochondrial genome of Paracondylactis sinensis, with 13 protein coding genes, one ORF, 2 tRNAs, and 2 rRNAs.
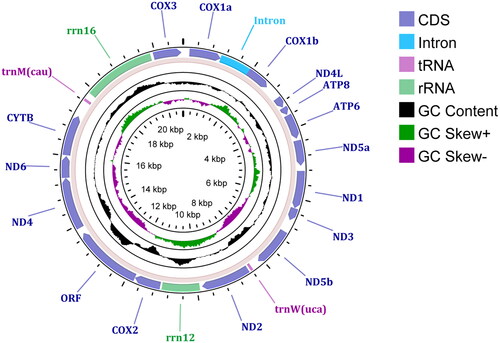
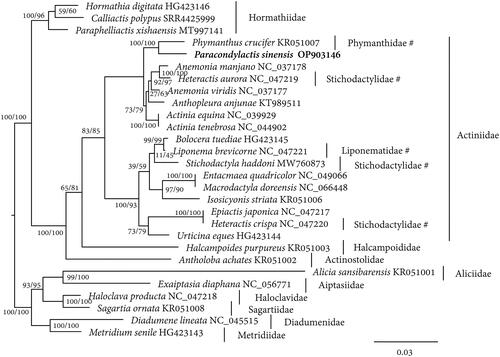
Figure 3. The maximum likelihood tree of Paracondylactis sinensis and other related sea anemones built from the concatenated amino acids of 13 protein coding genes. FLU + F + I + G4 model is used. Numbers near the nodes indicates SH-aLRT and ultrafast bootstrap support values from 20,000 replicates. The species P. sinensis whose sequence was produced in this study is in bold. The families each species belongs to are shown near the species name. The families that need further revision are marked with the symbols of hash.
Supplemental Material
Download TIFF Image (5.3 MB)Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OP903146. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA934839, SRR23454370, and SAMN33284299, respectively.
