Figures & data
Figure 1. Micranthes octopetala in its Native environment at Mt. Gwangdeoksan, Hwacheon-gun, Gangwon-do province, South Korea. The species reference image was taken by Sa-Bum Jang. M. octopetala is a perennial herb that grows to 15–30 cm and has 1–4 leaves. Flowers with 8 white petals bloom from June to July. Flowering stems are erect and leafless. The characteristic that can be distinguished from similar species (M. manchuriensis) is that it has an underground stolon.

Figure 2. The complete plastid genome of Micranthes octopetala. Graphic showing features of its plastome was generated using OGDraw. Colored boxes represent functional groups of plastid-coding genes. The clockwise and counter-clockwise transcribed genes are drawn inside and outside of the circle, respectively. The small gray bar graphs in the inner circle show the GC contents.
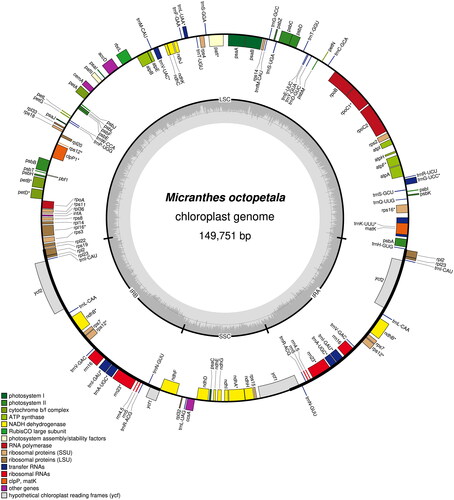
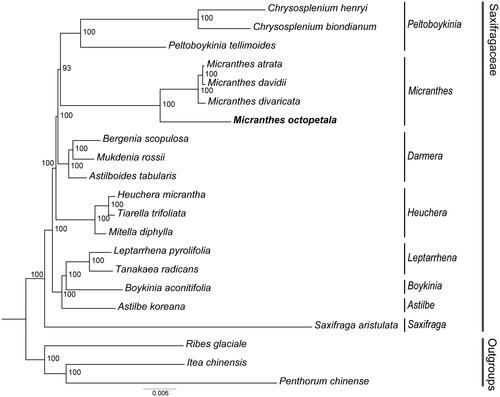
Figure 3. The ML tree of 18 species of Saxifragaceae based on concatenated protein-coding genes. M. octopetala is marked in bold. Bootstrap support values shown at the branches. The following sequences were used: Chrysosplenium henryi OK336532, Chrysosplenium biondianum OK336542, Peltoboykinia tellimoides MZ779205 (Yang et al. Citation2022), Micranthes atrata ON720899 (Yuan et al. Citation2023), Micranthes davidii ON720901 (Yuan et al. Citation2023), Micranthes divaricata ON720902 (Yuan et al. Citation2023), bergenia scopulosa KY412195 (Bai et al. Citation2018), mukdenia rossii MG470844 (Liu et al. Citation2018), astilboides tabularis MT316511 (Ha et al. Citation2020), heuchera micrantha OL489769, tiarella trifoliata MH708572, mitella diphylla MH708564, leptarrhena pyrolifolia MN496070 (Folk et al. Citation2020), tanakaea radicans MW300581 (Su et al. Citation2021), boykinia aconitifolia MN496058 (Folk et al. Citation2020), astilbe koreana MK990830, Saxifraga aristulata ON720851 (Yuan et al. Citation2023), Ribes glaciale MK887908, Itea chinensis MH191391 (Dong et al. Citation2018), and Penthorum chinense JX436155 (Dong et al. Citation2013).
Supplemental Material
Download JPEG Image (669.8 KB)Supplemental Material
Download JPEG Image (698.4 KB)Supplemental Material
Download JPEG Image (894.6 KB)Supplemental Material
Download MS Excel (12.3 KB)Data availability statement
The genome sequence data supporting the findings of this study are available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession no. OQ835312. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA956675, SRR24198730, and SAMN34227787, respectively.
