Figures & data
Figure 1. Adonis pseudoamurensis W.T. Wang 1980. This photo was taken by the author of this article, Li-Qiu Zhang. Herbs perennial, with two- to three-lobed leaves and flowers borne singly at the tips of stems or branches. Petals ca. 13, yellow; sepals purplish, about 1/2 as long as petals.

Figure 2. Chloroplast genome map of A. pseudoamurensis. Genes drawn outside the outer circle are transcribed counterclockwise, and genes drawn inside the outer circle are transcribed clockwise. Genes belonging to different functional groups are color-coded. The different colored legends in the bottom left corner indicate genes with different functions. The dark grey inner circle indicates the GC content of the chloroplast genome and the presence of nodes in the LSC, SSC, IR regions.
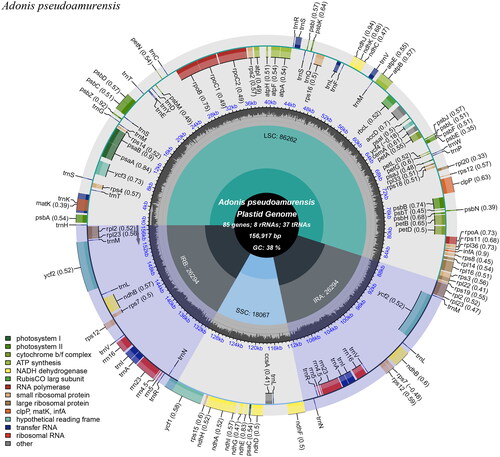
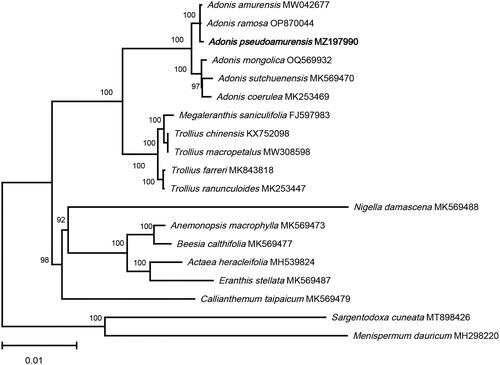
Figure 3. The ML phylogenetic tree is based on chloroplast genome sequences of Adonis pseudoamurensis species from the Ranunculaceae family, with Sargentodoxa cuneata (MT898426) and Menispermum dauricum (MH298220) as outgroups. Support values above the branches are ML bootstrap support. The following sequences were used: Trollius ranunculoides MK253447, Adonis coerulea MK253469 (He et al. Citation2019), Adonis pseudoamurensis MZ197990, Adonis ramosa OP870044, Megaleranthis saniculifolia FJ597983 (Kim et al. Citation2009), Trollius chinensis KX752098, Adonis sutchuenensis MK569470, Callianthemum taipaicum MK569479, Anemonopsis macrophylla MK569473, Beesia calthifolia MK569477, Eranthis stellata MK569487, Nigella damascena MK569488 (Zhai et al. Citation2019), Actaea heracleifolia MH539824 (Park et al. Citation2018), Menispermum dauricum MH298220 (Hina et al. Citation2018), Trollius farreri MK843818 (Yu et al. Citation2019), Adonis amurensis MW042677, Sargentodoxa cuneata MT898426 (Cui et al. Citation2021), Trollius macropetalus MW308598.
Supplemental Material
Download TIFF Image (131.3 KB)Supplemental Material
Download TIFF Image (289.1 KB)Supplemental Material
Download TIFF Image (262 KB)Supplemental Material
Download MS Word (635.1 KB)Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MZ197990. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA878453, SRR21533504, and SAMN30726550, respectively.
