Figures & data
Figure 1. Morphological characteristics of R. przewalskii subsp. przewalskii. The nutritional growth stage of R. przewalskii subsp. przewalskii, the leaf blade is leathery, often set at the end of the branch in an ovate-elliptic shape, and the apex is obtuse but with a small acumen. Its base is rounded or slightly heart-shaped, dark green above, glabrous, slightly wrinkled, midrib concave, lateral veins 11–12 pairs, retuse. In June 2022, the author took this photo in Xiazhasha Village, Xunhua County, Haidong City, Qinghai Province, China (102°44′8″E, 35°34′ 23″N, Altitude: 3499 m).

Figure 2. Annotated map of the circular chloroplast genome of R. przewalskii subsp. przewalskii. The genes inside the circles are transcribed clockwise and those outside are transcribed counterclockwise. The dark gray in the inner circle shows GC content, while the light gray shows a + T content (the dashed area in the inner circle indicates the content of the GC chloroplast genome). The legend identifies genes with different functions.
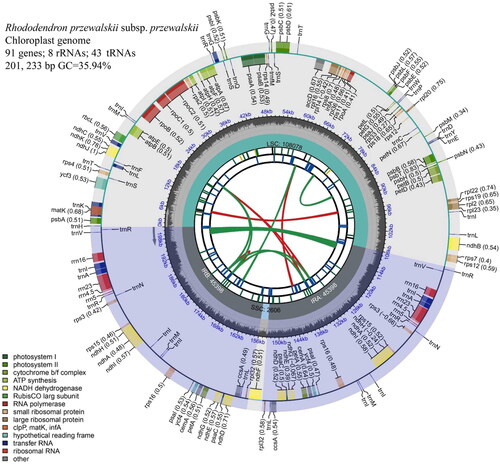
Figure 3. Maximum likelihood tree based on the chloroplast PCGs sequences of R. przewalskii subsp. przewalskii and 18 species of Ericaceae. The numbers shown next to the nodes are bootstrap support values and GenBank accession numbers of each species are listed in the tree. Red font for this study. Numbers >1 in the plot indicate support for species relatedness, and numbers <1 are bootstrap values, indicating node confidence levels.

Supplemental Material
Download MS Word (2.5 MB)Data availability statement
Supplementary Figure 2(A) is the sequencing depth map and coverage map and Supplementary Figure 2(B) is the black-white arrow shows the cis-spliced genes (left), and the black-grey-white arrow shows trans-spliced genes (rps12) (right), which the authors have placed in the Supplementary.
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI under the accession(https://www.ncbi.nlm.nih.gov/nuccore/OQ054014.1/). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA947480, SRR23938567, and SAMN33850058, respectively.
