Figures & data
Figure 1. Colony, appressorium morphology, and blast symptoms. (A) Mycelium growth on a petri dish (Photo taken by Jiaoyu Wang). (B) The three-celled conidia germinate and form an appressorium, the specialized penetration structure (Photo taken by Jiaoyu Wang). (C) Rice leaves with rice blast symptoms (Photo taken by Xiaohong Liu). scale bars are 1 cm, 10 µm, and 1 cm, respectively.
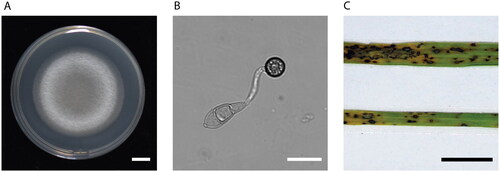
Figure 2. Genome map of Pyricularia oryzae Guy11 mitochondrion. This map was generated by CLC Genomics Workbench. Features were individually labeled.
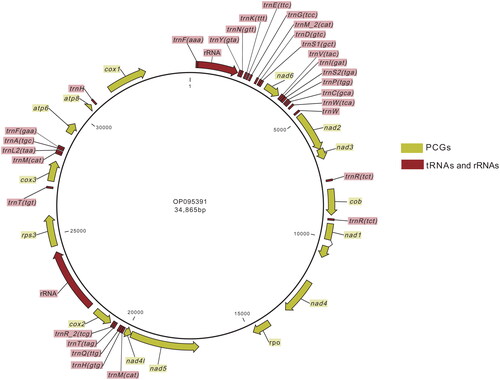
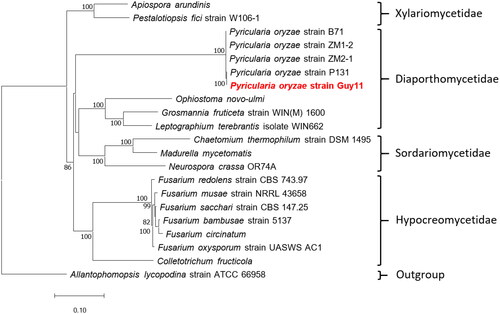
Figure 3. Phylogenetic tree of maximum likelihood (ML) method based on the mitogenome sequences of Pyricularia oryzae Guy11 and 19 Sordariomycetes species. The tree is rooted with Allantophomopsis lycopodina strain ATCC 66958 (Chen et al. Citation2023). bootstrap support values based on 1,000 replicates are displayed on each node as >70. The following sequences were used: Pyricularia oryzae strain ZM1-2 CM048866 (unpublished), Pyricularia oryzae strain B71 CP060338 (Peng et al. Citation2019), Pyricularia oryzae strain P131 CP114142 (unpublished), Pyricularia oryzae strain Guy11 OP095391 (this study), Pyricularia oryzae strain ZM2-1 CP099704 (unpublished), Allantophomopsis lycopodina strain ATCC 66958 CP103019 (unpublished), chaetomium thermophilum strain DSM 1495 JN007486 (Amlacher et al. Citation2011), madurella mycetomatis JQ015302 (van de Sande Citation2012), Fusarium circinatum JX910419 (Fourie et al. Citation2013), Neurospora crassa OR74A KC683708 (unpublished), Fusarium oxysporum strain UASWS AC1 KR952337 (unpublished), colletotrichum fructicola KX034082 (Liang et al. Citation2017), pestalotiopsis fici strain W106-1 KX870077 (Zhang et al. Citation2017), apiospora arundinis KY775582 (unpublished), ophiostoma novo-ulmi MG020143 (Abboud et al. Citation2018), Fusarium bambusae strain 5137 MH684411 (Wang et al. Citation2018), Fusarium redolens strain CBS 743.97 MT010909 (Yang et al. Citation2020), Fusarium sacchari strain CBS 147.25 MT010910 (Yang et al. Citation2020), Fusarium musae strain NRRL 43658 ON240982 (Degradi et al. Citation2022), leptographium terebrantis isolate WIN662 OP973818 (unpublished), grosmannia fruticeta strain WIN(M) 1600 OQ851465 (unpublished).
Supplemental Material
Download MS Word (138.5 KB)Supplemental Material
Download MS Word (635.2 KB)Supplemental Material
Download MS Word (5.6 MB)Supplemental Material
Download MS Word (16.2 KB)Data availability statement
The data supporting this study’s findings are openly available in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with accession number OP095391. The associated BioProject, BioSample, and SRA numbers are PRJNA866295, SAMN30160189/SAMN37189429, and SRR20852133/SRR20852132/SRR25783615.
