Figures & data
Figure 1. Photographs of Scutellaria barbata D. Don. (These photographs were taken by Prof. Quan Zhang). The foliage of S. barbata exhibits triangular, ovate, or ovate-lanceolate shapes, characterized by a sharp apex and a broad wedge-shaped or nearly truncated base. The raceme of this species is inconspicuous and positioned terminally. The lower bracts are elliptic or narrowly elliptic, while the bracteoles take the form of needle-shaped structures. Furthermore, the corollas of Scutellaria barbata display a vivid purple-blue coloration. (A) Plant panorama of S. barbata and (B) the flowers of S. barbata.

Figure 2. The circle map of chloroplast genome map of S. barbata. Distinctive colored boxes encircling the outer circle depict genes, with clockwise and counter-clockwise transcribed genes represented inside and outside the circle, respectively. The inner circle features a gray region indicating the GC content, while the quadripartite structure (LSC, SSC, IRA, and IRB) is illustrated on the inner circle accordingly.
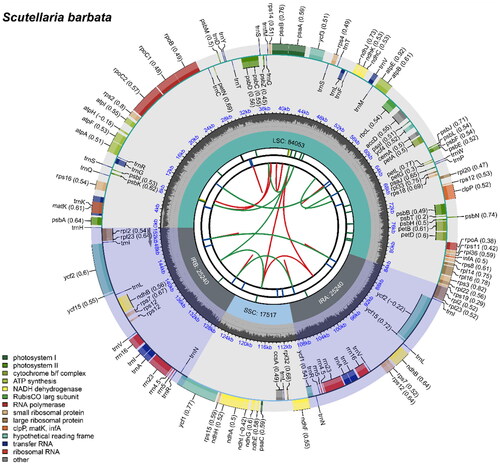
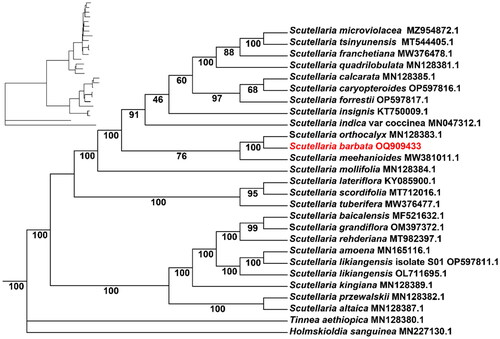
Figure 3. The maximum-likelihood phylogenetic tree of 25 Scutellaria species was constructed based on the CDS sequences extracted by IQ-TREE, with Tinnea aethiopica and Holmskioldia sanguinea added as outgroup. The phylogenetic tree was constructed using the maximum-likelihood method (ML) and bootstrap was performed 1000 times. The number on each branch indicates the boot support value. The following sequences were used: S. microviolacea MZ954872.1 (Wang et al. Citation2022), S. tsinyunensis MT544405.1 (Shan et al. Citation2021), S. franchetiana MW376478.1, S. calcarata MN128385.1 (Zhao et al. Citation2020), S. quadrilobulata MN128381.1 (Zhao et al. Citation2020), S. caryopteroides OP597816.1, S. forrestii OP597817.1, S. insignis KT750009.1, S. indica var coccinea MN047312.1 (Lee and Kim Citation2019), S. orthocalyx MN128383.1 (Zhao et al. Citation2020), S. meehanioides MW381011.1 (Zhang et al. Citation2021), S. mollifolia MN128384.1 (Zhao et al. Citation2020), S. lateriflora KY085900.1, S. scordifolia MT712016.1, S. tuberifera MW376477.1 (Shan et al. Citation2021), S. baicalensis MF521632.1 (Jiang et al. Citation2017), S. grandiflora OM397372.1, S. rehderiana MT982397.1, S. amoena MN165116.1 (Chen and Zhang Citation2019), S. likiangensis isolate S01 OP597811.1, S. likiangensis OL711695.1, S. kingiana MN128389.1 (Zhao et al. Citation2020), S. przewalskii MN128382.1 (Zhao et al. Citation2020), S. altaica MN128387.1 (Zhao et al. Citation2020), T. aethiopica MN128380.1 (Zhao et al. Citation2020), and H. sanguinea MN227130.1 (Lee and Kim Citation2020).
Supplemental Material
Download MS Word (198.8 KB)Data availability statement
The genome sequence data supporting the findings of this study can be publicly obtained at NCBI GenBank in https://www.ncbi.nlm.nih.gov with the accession number OQ909433. Associated BioProject, SRA, and Bio-Sample numbers are PRJNA947595, SRR23952789, and SAMN33861697.
